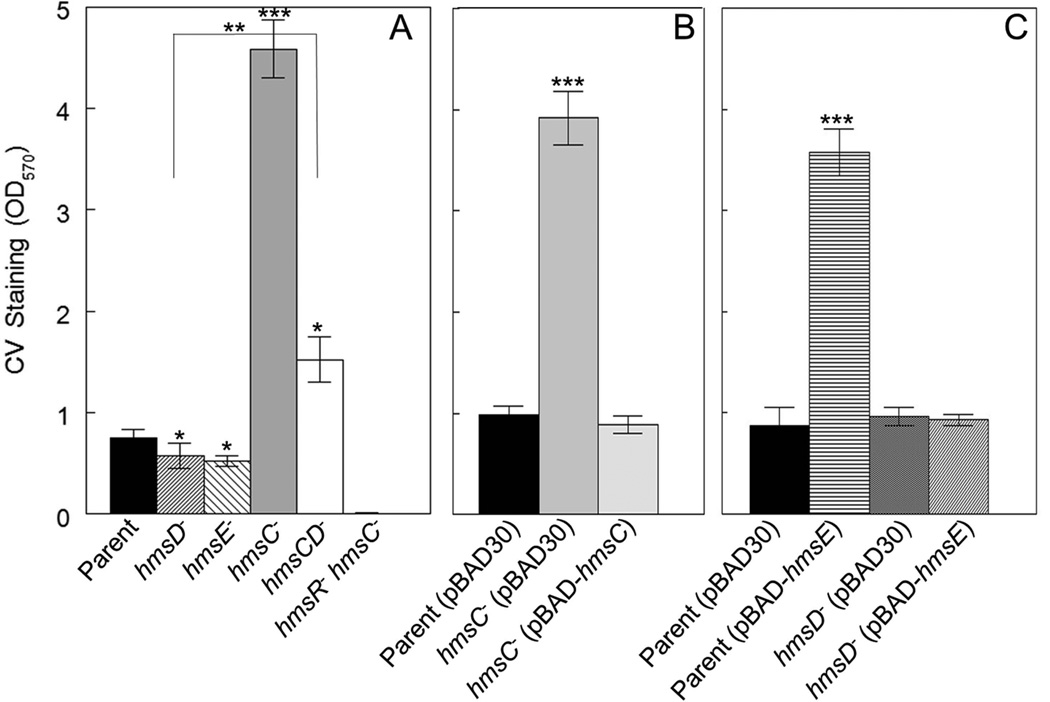
Fig. 1. HmsC and HmsE inversely modulate Hms-dependent biofilm formation in vitro via HmsD.
A crystal violet (CV) staining assay was used to assess Hms biofilm formation in the following strains: KIM6+ (Parent), KIM6-2159.1+ (hmsD−), KIM6-2174+ (hmsE−), KIM6-2173.1+ (hmsC−), KIM6-2198+ (hmsCD−) and KIM6-2173.2 (hmsC− hmsR−) (panel A). Panel B shows the results of complementation with hmsC expressed from plasmid pBAD-hmsC. Panel C demonstrates the effect of hmsE expressed from pBAD-hmsE on biofilm formation in the parent and hmsD mutant strains. Results are from duplicate assays on two independent cultures (4 biological samples) (in A and B) and from triplicate assays on three independent cultures (9 biological samples) (in C). Error bars indicate standard deviations. The asterisks indicate statistically significant differences between the parent strains and mutants by the Student’s T-test (*P<0.05, **P<0.005 *** P<0.001). The asterisks with the bracket indicate a statistically significant difference between the two indicated strains.