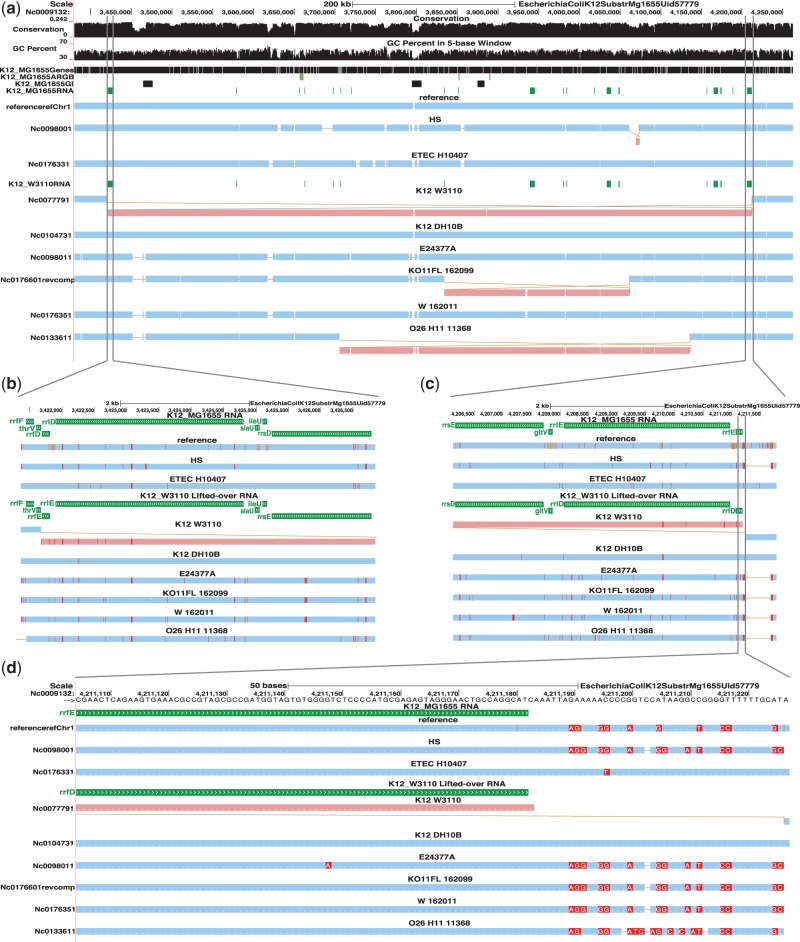
Fig. 1.
An example E.coli comparative assembly hub with E.coli K12 MG1655 as the reference browser. The top browser screenshot (a) shows an ∼900 kb region with a known large inversion (light red) in the closely related strain K12 W3110, which is flanked by homologous (with opposite orientations) ribosomal RNA operons rrnD and rrnE [Hayashi et al. (2006); Hill and Harnish (1981)], and is the result of recombination between them. (b–c) Zoom-in of the K12 W3110 inversion left and right boundaries, respectively, showing operon rrnE of K12 W3110 (‘K12_W3110 RNA’ track, in green, which is K12 W3110 ncRNA annotation track lifted-over to K12 MG1655) aligned to operon rrnD of K12 MG1655 (‘K12_MG1655 RNA’ track, also in green) on the left and operon rrnD of K12 W3110 aligned to operon rrnE of K12 MG1655 on the right. If further zoomed in (d), SNPs and query insertions are visible. The text on the screenshots was adjusted for better readability