Figure 1.
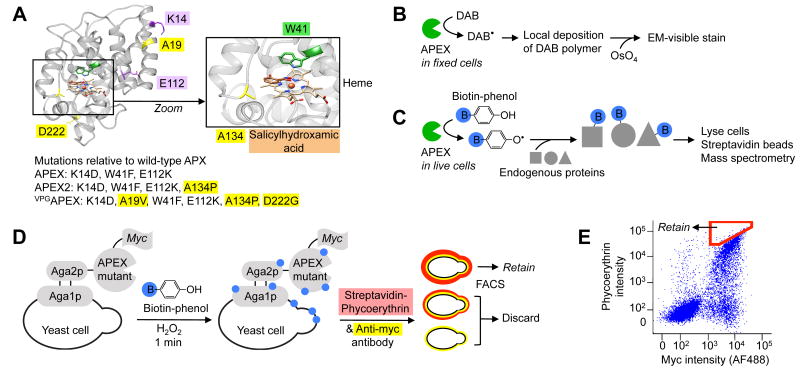
Yeast display evolution of APEX2 for electron microscopy (EM) and proteomics applications. (A) Structure of wild-type soybean ascorbate peroxidase (APX) with mutations present in APEX, APEX2, and VPGAPEX indicated. New mutations discovered in this study are highlighted yellow. The active site is magnified to show the heme cofactor, aromatic substrate (salicylhydroxamic acid, brown) binding site, and A134 position (yellow) that is mutated to proline in APEX2. From PDB ID 1 V0H9. (B) Scheme showing how APEX or APEX2 can be used as a reporter to generate contrast for EM.4 (C) Scheme showing how APEX or APEX2 can be used for proteomic tagging in living cells.5, 6 Blue B, biotin. (D) Labeling and selection scheme used to evolve APEX2 by yeast display. Biotin-phenol was added to a dilute yeast suspension for 1 minute to allow cells displaying highly active APEX variants to promiscuously biotinylate themselves. Minimal inter-cellular labeling was observed under these conditions. Biotinylation sites (blue) were stained with fluorescent streptavidin-phycoerythrin (red), and APEX expression level was quantified via anti-myc antibody staining (yellow). Two-dimensional fluorescence activated cell sorting (FACS) was used to enrich for cells displaying the highest activity/expression ratio (i.e., streptavidin/myc ratio). (E) FACS plot showing the initial APEX mutant library, and the sorting gate used in the first round of selection (red polygon). Single trial, 10,000 cells shown. FACS analyses of enriched populations and individual clones shown in Supplementary Figure 2.