Abstract
Purpose
The prostate cancer risk calculator from the Prostate Cancer Prevention Trial estimates the risk of positive biopsy and 1 containing high grade disease (Gleason score 7 or greater) based on prostate specific antigen, digital rectal examination, family history, race and prior negative biopsy. Since data used to create the calculator came from an unreferred population that underwent mainly sextant biopsy, to our knowledge its usefulness in the contemporary urology practice is unknown.
Materials and Methods
We performed the same multivariate logistic regression used to derive the prostate cancer risk calculator in a cohort of men from the Stanford Prostate Needle Biopsy Database who underwent initial prostate needle biopsy using an extended 12-core scheme.
Results
Our predictions of overall prostate cancer risk did not differ significantly from those of the calculator. Prostate specific antigen, abnormal digital rectal examination and family history were independent risk factors. However, our model predicted a much greater risk of high grade disease than the prostate cancer risk calculator. Prostate specific antigen, abnormal digital rectal examination and age were independent risk factors for high grade disease.
Conclusions
The difference between our estimated risk of high grade prostate cancer and that of the prostate cancer risk calculator can be potentially explained by 1) differences between the cohorts (referred vs unreferred) or 2) the difference in grading, ie grading accuracy due to the difference in biopsy schemes or to temporally related grade shifts. Caution should be used when applying the prostate cancer risk calculator to counsel patients referred for suspicion of prostate cancer since it underestimates the risk of high grade disease.
Keywords: prostate, prostatic neoplasms, risk, nomograms, prognosis
Prostate cancer has the highest incidence of all cancers affecting men in the United States, accounting for 25% of newly diagnosed malignancies. At 27,360 deaths per year, it is the second leading cause of cancer related death.1 With the advent of screening for prostate cancer using PSA the estimated lifetime risk of prostate cancer has increased from 9% to 15.8%.2 Although its use as a screening tool for prostate cancer remains controversial,3-5 PSA is nevertheless a sensitive prostate cancer indicator that has been traditionally used by clinicians to estimate prostate cancer risk and counsel patients accordingly in conjunction with clinical parameters such as age, family history, DRE and race. Until recently no tool has been available to objectively quantify the risk of prostate cancer in a given patient based on his known risk factors.
PCPT was a landmark prospective, randomized, placebo controlled trial with the primary end point of a 24.8% decrease in prostate cancer risk in men on finasteride vs placebo.6 The trial enrolled 18,882 healthy men 55 years old or older with PSA 3.0 ng/ml or less and normal DRE, and randomized them to placebo or finasteride. Participants underwent prostate needle biopsy when clinically indicated due to abnormal DRE or increasing PSA, or at the end of the study. In secondary analysis of the placebo arm of the trial the investigators noted that prostate cancer developed at all PSA ranges, even those well below the traditional 4.0 ng/ml cutoff that would typically prompt biopsy. This cohort consequently became a convenient population in which to study prostate cancer risk factors.
Post hoc analysis using multivariate logistic regression was used to model the risk of overall and high grade prostate cancer, defined as Gleason score 7 or greater, at biopsy. In this model PSA, family history of prostate cancer and abnormal DRE were independent positive risk factors for positive biopsy at any Gleason score. PSA, abnormal DRE, black race and increasing age were independent positive risk factors for positive biopsy containing high grade disease.7 Ultimately the risk equations from this model were used to develop PCRC.8 This nomogram predicts the risk of positive biopsy or positive biopsy containing high grade disease using the previously identified independent risk factors. It is available on the Internet as a tool for clinicians to assess prostate cancer risk in their patients and counsel them on the likelihood of positive biopsy results.
Unfortunately more than 80% of biopsies done during PCPT used the traditional sextant scheme, bringing into question the usefulness of this nomogram in the era of extended biopsy schemes. Also, since PCRC was derived in a cohort of essentially healthy men with low PSA who were then followed longitudinally, the appropriateness of using this calculator in a urological practice where patients routinely have increased PSA and often multiple risk factors for prostate cancer has also been questioned. Despite these limitations 2 recent studies validated PCRC in modern cohorts in referral populations undergoing extended biopsy, although there is 1 conflicting report.9-11 However, these studies did not specifically examine the ability of PCRC to accurately predict the risk of high grade disease, nor was that their intent.
To address these concerns we identified a cohort of patients referred to our institution for suspicion of prostate cancer (abnormal PSA or abnormal DRE) who subsequently underwent initial prostate needle biopsy using an extended 12-core scheme. We modeled the risk of positive biopsy and the risk of high grade disease using the same statistical methods described by the PCRC developers.
MATERIALS AND METHODS
Study Cohort
SPNBD is a prospectively maintained database of all biopsies done in our practice. In this database we retrospectively identified 636 men who underwent initial biopsy using a 12-core extended biopsy scheme from 1999 to 2004. Some patients who were missing values for the independent risk factors identified in the original PCRC risk models were excluded from each model, as described. Since all patients underwent initial biopsy, our analysis did not include previous biopsy as a putative risk factor. However, the absence of previous biopsy in our model did not interfere with our ability to compare predictions in men undergoing initial biopsy. Also, although our study group was more ethnically diverse, only black race was analyzed as a potential risk factor to achieve parity with the original PCRC. All pathological results were reviewed by a single pathologist.
Prostate Cancer Risk
We performed complete case analysis, omitting 17 men (2.7%) with missing PSA or DRE result in the data set. We performed multivariate logistic regression of disease status (positive vs negative biopsy) on the natural logarithm of PSA in ng/ml, family history (yes vs no) and DRE result (normal vs abnormal). We used the Wald test to determine the significance of each independent variable, controlling for the other 2. To test the equality of each coefficient to the corresponding 1 from PCRC we performed the z test on the difference, inferring the SE of PCRC model coefficients from the reported CIs.
High Grade Cancer Risk
We adopted the PCRC definition of high grade prostate cancer as that with a Gleason score of 7 or greater. We performed similar complete case analysis, omitting 81 men (12.7%) with missing PSA, DRE result or ethnicity in the data set. We performed multivariate logistic regression of high grade disease status (high vs low grade vs no cancer) on the natural logarithm of PSA in ng/ml, patient age at biopsy in years, DRE result (normal vs abnormal) and ethnicity (black vs nonblack).
RESULTS
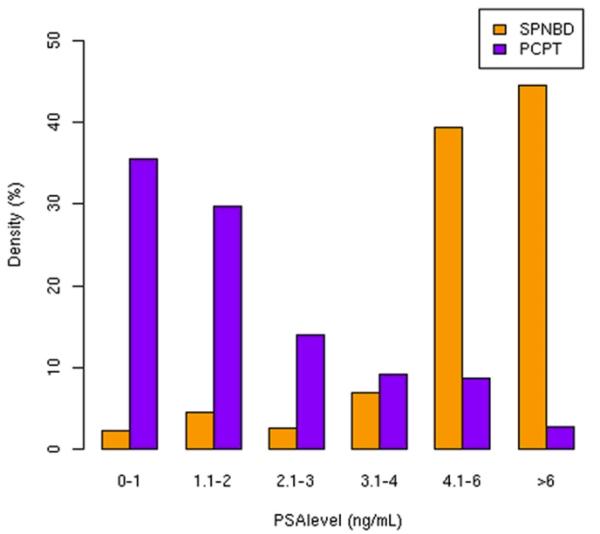
Table 1 lists the clinical characteristics of the 636 men in the Stanford cohort compared with the 5,519 in the PCRC analysis. A significant number of men in the Stanford cohort (20.3%) were younger than 55 years old, a group excluded from PCPT. In contrast, almost half of the patients in the PCRC sample were older than 70 years old. The racial profile of our group was also more diverse than in PCPT with 3.8% of the men identified as Hispanic and 10.1% identified as Asian. The rates of black men were similar. The prevalence of a positive family history of prostate cancer was similar in the 2 cohorts. As expected in a referral population, the SPNBD cohort had much higher PSA than the PCRC sample (median 5.7 vs 1.5 ng/ml) (fig. 1).
Table 1. Characteristics of Stanford study cohort and PCRC analysis sample.
| Characteristic | No. Stanford (%) | No. PCRC (%) |
|---|---|---|
| Age: | ||
| Less than 40 | 3 (0.5) | — |
| 40–44 | 7 (1.1) | — |
| 45–49 | 25 (3.9) | — |
| 50–54 | 94 (14.8) | — |
| 55–59 | 118 (18.6) | 38 (0.7) |
| 60–64 | 123 (19.3) | 1,143 (20.7) |
| 65–69 | 118 (18.6) | 1,741 (31.5) |
| Greater than 70 | 148 (23.3) | 2,597 (47.1) |
| Prostate Ca family history: | ||
| No | 513 (80.7) | 4,599 (83.3) |
| Yes | 123 (19.3) | 920 (16.7) |
| Race: | ||
| White | 456 (71.7) | 5,276 (95.6) |
| Black | 24 (3.8) | 175 (3.2) |
| Asian | 64 (10.1) | — |
| Hispanic | 24 (3.8) | — |
| Other/unknown | 68 (10.7) | 68 (1.2) |
| Prior biopsy: | ||
| None | 636 (100) | 4,873 (88.3) |
| 1 or More | — | 646 (11.7) |
| DRE: | ||
| Normal | 440 (69.3) | 4,968 (90.0) |
| Abnormal | 196 (30.7) | 551 (10.0) |
Figure 1.
In most patients in PCRC analysis cohort PSA was between 0 and 2 ng/ml but in Stanford cohort PSA was greater than 4 ng/ml.
Table 2 lists the absolute number and proportion of men with prostate cancer and high grade prostate cancer, stratified by PSA. In the Stanford cohort the percent of men with cancer was double that in the PCRC sample and the percent with high grade disease was 7-fold higher.
Table 2. Patients with identified and high grade prostate cancer by PSA.
| All Grades Prostate Ca |
High Grade Prostate Ca |
||||
|---|---|---|---|---|---|
| PSA (ng/ml) | No. Stanford Pts |
No. Stanford (%) |
% PCRC | No. Stanford (%) |
% PCRC |
| 0–1 | 14 | 1 (7.1) | 11.1 | 1 (7.1) | 1.0 |
| 1.1–2 | 28 | 9 (32.1) | 20.5 | 7 (25.0) | 2.6 |
| 2.1–3 | 16 | 8 (50.0) | 26.5 | 4 (25.0) | 5.7 |
| 3.1–4 | 43 | 18 (41.9) | 30.0 | 12 (27.9) | 9.4 |
| 4.1–6 | 246 | 97 (39.4) | 48.6 | 65 (26.4) | 4.6 |
| Greater than 6 | 278 | 158 (56.8) | 43.3 | 129 (46.4) | 22.0 |
| Overall | 625 | 291 (46.6) | 21.9 | 218 (34.9) | 4.7 |
Table 3 shows baseline odds and ORs in the 2 models. The ORs of log(PSA) (corresponding to a 1 ng/ml increase in PSA), abnormal DRE and positive family history were significant for overall prostate cancer detection since in each the 95% CI excluded 1. For high grade prostate cancer PSA, abnormal DRE and age were significant but black race was not.
Table 3. Stanford model and PCRC ORs.
| Prostate Ca Risk Factor | Stanford Point Estimate (95% CI) |
PCRC Point Estimate (95% CI) |
||
|---|---|---|---|---|
| Overall: | ||||
| Intercept (baseline odds) | 0.12 | (0.07–0.22) | 0.17 | (0.15–0.18) |
| Log(PSA) | 2.43 | (1.84–3.21) | 2.34 | (2.14–2.56) |
| DRE | 2.55 | (1.74–3.73) | 2.48 | (2.03–3.00) |
| Family history | 1.69 | (1.10–2.60) | 1.31 | (1.11–1.55) |
| High grade: | ||||
| Intercept (baseline odds) | 0.004 | (0.001–0.015) | 0.002 | (0.000–0.010) |
| Log(PSA) | 2.22 | (1.61–3.08) | 3.63 | (3.03–4.35) |
| DRE | 3.03 | (1.97–4.65) | 2.72 | (1.95–3.78) |
| Age | 1.05 | (1.03–1.08) | 1.03 | (1.01–1.05) |
| Black | 2.12 | (0.84–5.35) | 2.61 | (1.55–4.39) |
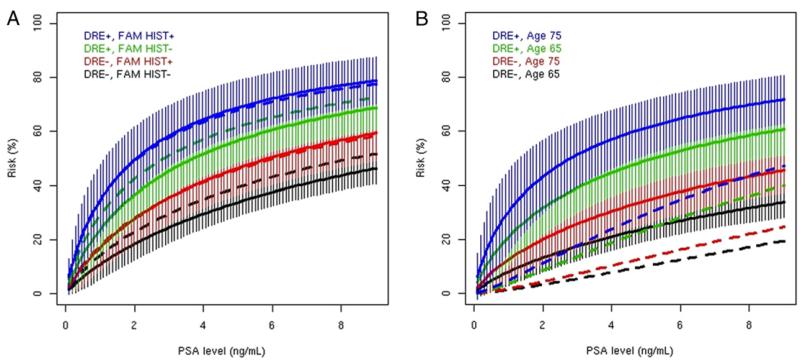
In the overall prostate cancer model based on the z test none of the coefficients differed significantly from the corresponding coefficient in the PCRC model. When graphed, the PCRC risk predictions were within the error bars of our model (fig. 2, A). For high grade prostate cancer the PCRC risk predictions were well below our error bars (fig. 2, B). The only statistically significantly different coefficient was that of log(PSA), which was smaller in our model (z test statistic −2.58, p = 0.01).
Figure 2.
PCRC cohort curves (broken lines) of prostate cancer risk fall within those of Stanford cohort curve (solid lines) error bars. A, overall. B, high grade. FAM HIST, family history.
ROC curves using the PCRC model on our data set for overall and high grade prostate cancer resulted in an AUC of 0.664 and 0.509, respectively. Corresponding values in PCPT were 0.702 and 0.698, respectively.7
DISCUSSION
Referrals for men suspected of having prostate cancer make up a substantial proportion of new patients seen in a urological practice. Historically recommendations on whether to perform prostate needle biopsy have been based on subjective inferences about individual risk factors for prostate cancer weighed against the potential morbidity of the procedure and the risk of over detection (the risk of finding indolent cancer that, if detected and treated, would be considered unnecessary treatment). Since a tool to quantify the patient risk of prostate cancer was not available, the threshold for recommending biopsy has traditionally been low and variable among providers. In patients whom most clinicians would deem at low risk, there was no optimal way to answer the often asked question of what represented low. Conversely in patients at high risk in whom almost every clinician would recommend biopsy it was difficult to estimate of the odds of positive biopsy in any way that was supported by empirical data.
The PCRC from the PCPT was 1 of the first models of prostate cancer risk derived in a large cohort. Although the data that underlie PCRC were accrued more than a decade ago, it remains in use today by a wide range of individuals, including patients, primary care physicians and urologists. Since most biopsies done in PCPT were sextant, the validity of PCRC predictions in contemporary patients, of whom most undergo 12-core biopsy, is unclear. Also, some groups have questioned the generalizability of the PCRC to patients typically seen in modern urological practice. Eligibility for PCPT included PSA 3.0 ng/ml or less, normal DRE and age 55 years or greater (50 years in black men). Two separate large series have validated PCRC use in modern urological practice. However, the largest series, which was recently published, shows statistically significant differences between coefficients in the risk equations, suggesting that the PCPT model does not calibrate well with those derived from more modern data sets.9,10,11
In the SPNBD analysis using the statistical methods described by the PCRC investigators there were no statistically significant differences between the risk estimates for overall prostate cancer detection. Since extended biopsy leads to a higher cancer detection rate,12 one would expect a model derived from contemporary 12-core data to predict higher risk, although as noted only 1 of 3 current studies confirmed this hypothesis. The Stanford data do not support this supposition. A possibility to explain this observation relates to differences in the study populations. Median PSA in the PCPT placebo arm was 1.5 ng/ml while median PSA in the Stanford cohort was 5.7 ng/ml. Our study population may have been enriched in patients with benign prostatic hyperplasia. This inverse relationship between prostate size and the cancer detection rate is well established.13
Analysis of SPNBD yielded larger estimates of the risk of high grade prostate cancer compared to those of PCRC. That is, when controlling for PSA, DRE, age and race, the model based on SPNBD predicted a higher risk of high grade disease. The difference may have been due in part to the difference between the cohorts. SPNBD consisted of men referred to our institution for suspicion of prostate cancer while PCPT accrued men without suspicion of prostate cancer. To join PCPT patients had normal DRE and PSA less than 3 ng/ml. They later underwent biopsy if PSA or DRE became abnormal or they attained the study end. With respect to PSA changes prompting biopsy one assumes that the absolute magnitude of the increase was relatively small (PSA had to exceed 4 ng/ml). In a referral population the absolute magnitude of PSA increases may be higher.
A second possible explanation for the difference in estimates is the difference in grading. We previously reported that traditional sextant biopsy under graded 1/4 tumors in radical prostatectomy specimens.14 However, based on this observation alone it would be difficult to explain the vast difference between the 2 models for high risk prostate cancer. Another explanation of these differences may be attributable to pathological grade inflation, which is a well described phenomenon that has led to contemporary Gleason scores being about 1 point higher compared to those in historical controls.15 Thus, while the prostate cancer detection rate has remained essentially the same, a greater proportion of biopsies are being assigned a higher Gleason score. However, all of our biopsies were read by a single pathologist considered to be an expert in prostate pathology.
Another 2 groups validated PCRC for overall prostate cancer but they did not explicitly examine the risk of biopsy with high grade disease.9,10 A third study showed weaker associations for overall and high grade cancer detection (AUC = 0.57 and 0.60, respectively).
PCRC is an important, widely used tool that estimates prostate cancer risk. Unfortunately it underestimates the risk of high grade disease in the subset of men referred to urologists for suspicion of prostate cancer, who are those most likely to use and benefit from the calculator. This finding highlights the pitfall of attempting to generalize a model derived in a cohort with characteristics different than the target population. Artificially low estimates of the risk of high grade disease may inappropriately deter patients from undergoing prostate needle biopsy or delay a primary care physician from referring a patient for urological evaluation.
PCRC performs better than PSA alone to predict prostate cancer (AUC 0.691 vs 0.655, p = 0.009).9 However, the model is far from perfect and may benefit from including other variables not available at the time of its conception. Notably ethnicity information in PCPT was almost entirely limited to white and black men, who comprised 98.8% of the study population. Potential genetic markers in various ethnic populations have recently surfaced that confer an increased risk of prostate cancer and high grade prostate cancer.16 Future studies should be directed toward this end. In conclusion, caution should be used when applying PCRC risk estimates in patients seen in urological practice since it under-estimates the risk of high grade disease.
ACKNOWLEDGMENTS
Dr. John McNeal reviewed pathological results.
Study received institutional review board approval.
Supported by Clinical and Translational Science Award UL1 RR025744 and Cancer Center Support Grant P30 CA124435 (BBT, PWL).
Abbreviations and Acronyms
- DRE
digital rectal examination
- PCPT
Prostate Cancer Prevention Trial
- PCRC
Prostate Cancer Risk Calculator
- PSA
prostate specific antigen
- SPNBD
Stanford Prostate Needle Biopsy Database
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer Statisitics, 2009. CA Cancer J Clin. 2009;59:225. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Seidzman Mushinski MH, Gelb SK, et al. Probabilities of eventually developing or dying of cancer—United States. CA Cancer J Clin. 1985;35:36. doi: 10.3322/canjclin.35.1.36. [DOI] [PubMed] [Google Scholar]
- 3.Concato J, Wells CK, Horwtiz RI, et al. The effectiveness of screening for prostate cancer: a nested case-control study. Arch Intern Med. 2006;166:38. doi: 10.1001/archinte.166.1.38. [DOI] [PubMed] [Google Scholar]
- 4.Andriole GL, Crawford ED, Grubb RL, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schröder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 6.Thomspon IM, Goodman PJ, Tangen CM, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003;349:215. doi: 10.1056/NEJMoa030660. [DOI] [PubMed] [Google Scholar]
- 7.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 8. [Accessed October 1, 2009];Risk of Biopsy-Detectable Prostate Cancer. Available at http://deb.uthscsa.edu/URORiskCalc/Pages/uroriskcalc.jsp.
- 9.Eyre SJ, Ankerst DP, Wei JT, et al. Validation in a multiple urology practice cohort of the Prostate Cancer Prevention Trial calculator for predicting prostate cancer detection. J Urol. 2009;182:2653. doi: 10.1016/j.juro.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez DJ, Han M, Humphreys EB, et al. Predicting the outcome of prostate biopsy: comparison of a novel logistic regression-based model, the prostate cancer risk calculator, and prostate-specific antigen level alone. BJU Int. 2009;103:609. doi: 10.1111/j.1464-410X.2008.08127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen CT, Yu C, Moussa A, et al. Performance of Prostate Cancer Prevention Trial risk calculator in a contemporary cohort screened for prostate cancer and diagnosed by extended prostate biopsy. J Urol. 2010;183:529. doi: 10.1016/j.juro.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Matlaga BR, Eskew LA, McCullough DL. Prostate biopsy: indications and technique. J Urol. 2003;169:12. doi: 10.1016/S0022-5347(05)64024-4. [DOI] [PubMed] [Google Scholar]
- 13.Uzzo RG, Wei JT, Waldbaum RS, et al. The influence of prostate size on cancer detection. Urology. 1995;46:831. doi: 10.1016/s0090-4295(99)80353-7. [DOI] [PubMed] [Google Scholar]
- 14.King KR, McNeal JE, Gill H, et al. Extended prostate biopsy scheme improves reliability of Gleason grading: implications for radiotherapy patients. Int J Radiat Oncol Biol Phys. 2004;59:386. doi: 10.1016/j.ijrobp.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Albertsen PC, Hanley JA, Barrows GH, et al. Prostate cancer and the Will Rogers phenomenon. J Natl Cancer Inst. 2005;97:1248. doi: 10.1093/jnci/dji248. [DOI] [PubMed] [Google Scholar]
- 16.Chen M, Huang YC, Yang S, et al. Common variants at 8q24 are associated with prostate cancer risk in Taiwanese men. Prostate. 2010;80:502. doi: 10.1002/pros.21084. [DOI] [PubMed] [Google Scholar]