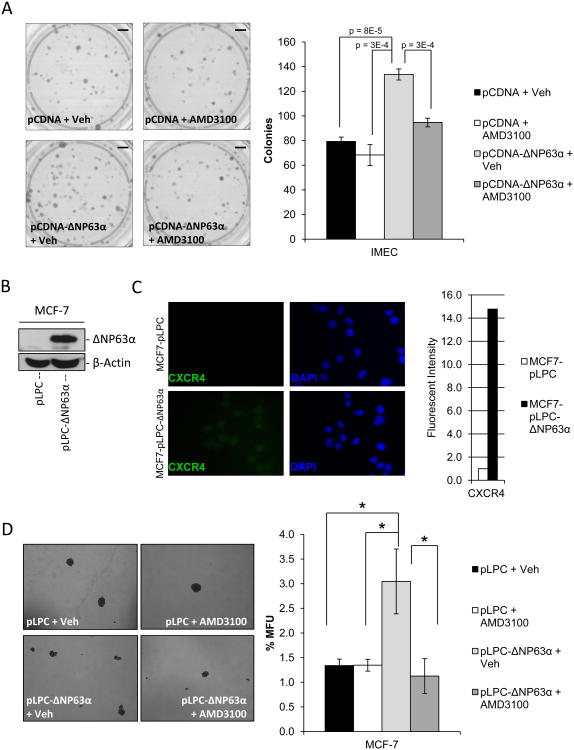
Figure 3. ΔNP63α-dependent regulation of stem cell activity is mediated through induction of CXCR4 expression.
A) Clonogenicity assays were performed in SUM-102 cells that were transfected with either linearized pCDNA or pCDNA-ΔNP63α. Cells were then plated at clonogenic capacity in the presence or absence of the CXCR4 antagonist AMD3100 (5 μM). Images of colonies stained with crystal violet (left), quantification of the number of colonies formed per experimental group (right). P-values are indicated. Scale bars, 850 mm. B) Western immunoblot to detect ΔNP63α protein expression in protein lysates of MCF-7 cells stably infected with an empty retrovirus (MCF7-pLPC) or one encoding ΔNP63α (MCF7-pLPC-ΔNP63α). C) Representative immunofluorescence images of MCF7-pLPC+/- ΔNP63α cells to detect CXCR4 protein expression levels (left), quantification of fluorescent intensity of image normalized to cell number (fluorescent intensity of DAPI staining, right) 40× magnification. D) MCF7-pLPC+/- ΔNP63α were seeded on non-adherent tissue culture plates at 500 cells/mL in the presence or absence of the CXCR4 antagonist AMD3100 (5 μM). Mammosphere forming capacity was assessed 6 days post-plating. Representative 10× phase images of mammospheres (left), quantification of mammosphere forming units (MFU) (right). All data are mean ± S.D.; n=3, asterisks indicates a P<0.013.