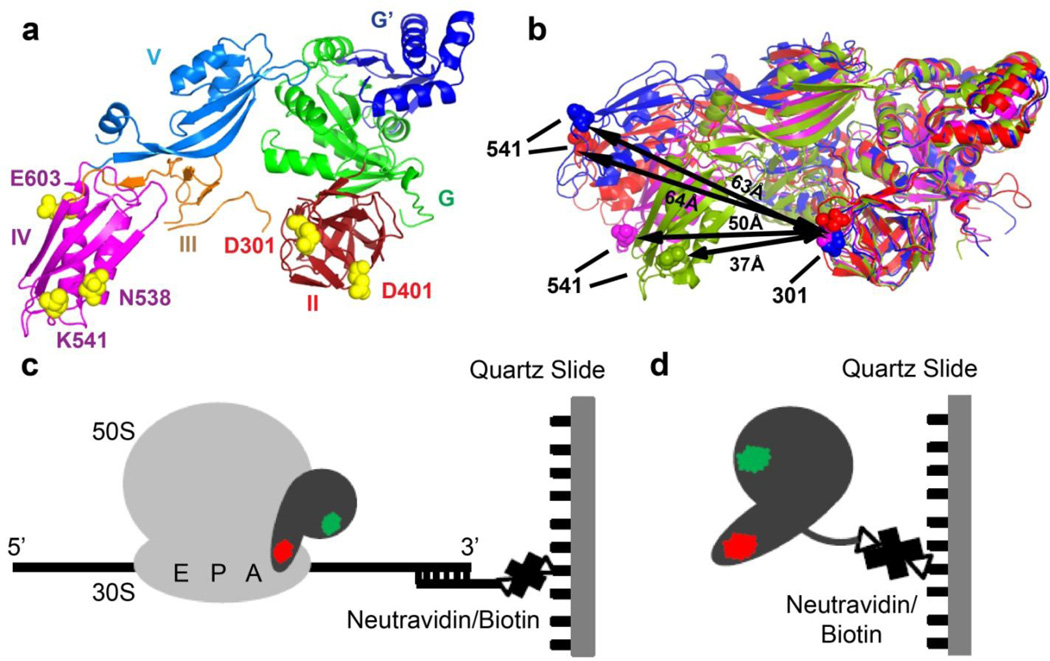
FIGURE 1. Experimental design.
(a) Residues chosen for fluorescent labeling (yellow spheres, E. coli numbering) viewed in the crystal structure of ribosome-free EF-G from T. thermophilus (PDB code 1dar12). Domains of EF-G are colored as shown. (b) Comparison of conformations in ribosome-bound and ribosome-free EF-G: EF-G (T. thermophilus) bound to the posttranslocation ribosome is shown in blue (PDB code 2wri16); EF-G (E. coli) bound to the pretranslocation ribosome in the presence of viomycin is shown in red (PDB code 3j5×5); ribosome-free EF-G•GDP (T. thermophilus) is shown in magenta (PDB code 1dar12) and ribosome-free EF-G (S. aureus) is shown in green (PDB code 2×ex17). Domains I and II of the EF-G structures were superimposed. Distance between the α-carbon atoms of residues 541 and 301 (E.coli numbering, shown as spheres) used for fluorescent labeling in respective structures is indicated by black arrows. (c) During smFRET imaging, EF-G doubly-labeled with donor (green) and acceptor (red) dyes was bound to ribosomes, which were immobilized by hybridization of the mRNA 3’ tail to a biotin-derivatized DNA oligonucleotide tethered via neutravidin to a microscope slide. (d) In smFRET studies of ribosome-free EF-G, EF-G doubly-labeled with donor (green) and acceptor (red) dyes was immobilized via C-terminal biotinylated-tag.