Abstract
To assess the impact of the genetic variation in toll-like receptors (TLR) on outcome after allogeneic myeloablative conditioning hematopoietic cell transplantation (HCT) we have investigated 29 single nucleotide polymorphisms (SNP) across 10 TLRs in 816 patients and donors. Only donor genotype of TLR8 rs3764879, which is located on the X chromosome, was significantly associated with outcome at the Bonferroni corrected level P≤0.001. Male hemizygosity and female homozygosity for the minor allele were significantly associated with disease free survival (DFS) (hazard ratio (HR) 1.47 (95% confidence interval (CI) 1.16–1.85); P=0.001). Further analysis stratified by donor sex due to confounding by sex, was suggestive for associations with overall survival (male donor: HR 1.41 (95% CI 1.09–1.83), P=0.010); female donor: (HR 2.78 (95% CI 1.43–5.41), P=0.003), DFS (male donor: HR 1.45 (95% CI 1.12–1.87), P=0.005; female donor: HR 2.34 (95% CI 1.18–4.65), P=0.015) and treatment related mortality (male donor: HR 1.49 (95% CI 1.09–2.04), P=0.012; female donor: HR 3.12 (95% CI 1.44–6.74), P=0.004).
In conclusion our findings suggest that the minor allele of TLR8 rs3764879 of the donor is associated with outcome after myeloablative conditioned allogeneic HCT.
Keywords: Toll-like receptors, Toll-like receptor 8, Allogeneic hematopoietic cell transplantation, single nucleotide polymorphism
INTRODUCTION
After allogeneic hematopoietic cell transplantation (HCT), both innate and adaptive immune mechanisms are involved in the immune reactions that result in graft versus host disease (GVHD) and the curative graft versus tumor effect (GVT) (1;2). Although the importance of human leukocyte antigen (HLA) matching on outcome after allogeneic HCT is well recognized (3), the significance of genetic variation in immune response genes outside the major histocompatibility complex system has become increasingly evident (4–8).
The toll-like receptors (TLR) are germline encoded pattern recognition receptors which recognize specific microbial pathogen associated molecular patterns (PAMP) and endogenous alarmins (9–12). TLRs are mainly expressed on antigen presenting cells, and play a central role in immune surveillance and in the initiation of the inflammatory response (9;13). Ten different TLR (TLR1–10) have been identified in humans. After allogeneic HCT TLRs may not only be involved in the immune response to infections, but also in acute GVHD, where tissue damage induced by high dose conditioning may lead to a milieu with ample supply of TLR ligands (14;15). Furthermore, TLRs are thought to be involved in GVT, as administration of TLR agonists has been shown to induce antitumor immunity and tumor regression in in-vivo tumor models and clinical trials (16–19).
In the setting of allogeneic HCT the TLR genes have been studied in terms of the impact of their variation on outcome and susceptibility to infection. Although single nucleotide polymorphisms (SNPs) in several TLR genes have been investigated, the most extensively studied are two functional SNPs in TLR4, Thr399Ile and Asp299Gly. These SNPs have been associated with acute GVHD, invasive aspergillosis and hemorrhagic cystitis (20–23).
The objective of the current study is to investigate associations between 29 SNPs across 10 TLR genes (Supplemental table S1) and outcome in a cohort of 816 patients and donors undergoing myeloablative conditioning matched unrelated donor allogeneic HCT for advanced hematological malignancies.
PATIENTS AND METHODS
The study cohort consisted of 816 donor/recipient pairs with acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML) or myelodysplastic syndrome (MDS) undergoing myeloablative hematopoietic cell transplantation with bone marrow or granulocyte colony stimulating factor (G-CSF) mobilized peripheral blood stem cells (PBSC) from 10/10 allele (HLA-A, B, C, DRB1 and DQB1) matched unrelated donors. Early stage disease was defined as AML and ALL in first complete remission, CML in first chronic phase and MDS with refractory anemia with or without ringed sideroblasts. Intermediate stage disease was defined as AML and ALL in second or subsequent complete remission or in first relapse, CML in accelerated phase or greater than first chronic phase. Advanced stage disease was defined as AML or ALL in second or subsequent relapse or primary induction failure, and CML in blast phase, MDS subtype refractory anemia with excess blasts or in transformation, or MDS not otherwise specified. Transplantation demographics are shown in table 1. The median follow-up was 11.1 (range 0.8–22) years.
Table 1.
Transplantation demographics
| Variable | |
|---|---|
| Number of patients, no. | 816 |
| Number of centers, no. | 89 |
| Male patient gender | 470 (58) |
| Patient age, median (range), years | 37 (<1–65) |
| 0–9 years | 61 (7) |
| 10–19 years | 75 (9) |
| 20–29 years | 130 (16) |
| 30–39 years | 206 (25) |
| 40–49 years | 236 (29) |
| ≥50 years | 108 (13) |
| Donor age, median (range), years | 36 (18–59) |
| Sex of donor/patient | |
| Female/male | 144 (18) |
| Other combinations | 672 (82) |
| Karnofsky prior to transplant ≥ 90 (only evaluable for 789 patients) | 627 (79) |
| CMV serostatus of donor/patient | |
| Negative/negative | 328 (40) |
| Other combinations | 461 (56) |
| unknown | 27 (3) |
| Donor previous pregnancies (female only, n=346) | |
| 0 | 118 (34) |
| 1 | 50 (14) |
| ≥2 | 116 (34) |
| unknown | 62 (18) |
| Disease at transplant | |
| Acute myeloid leukemia | 126 (15) |
| Acute lymphoblastic leukemia | 138 (17) |
| Chronic myeloid leukemia | 390 (48) |
| Myelodysplastic syndrome | 162 (20) |
| Disease stage at transplant | |
| Early | 651 (80) |
| Intermediary | 72 (9) |
| Advanced | 85 (10) |
| Other | 8 (1) |
| Graft Type | |
| Bone marrow | 711 (87) |
| Peripheral blood stem cells | 105 (13) |
| Graft-versus-host disease prophylaxis | |
| Tacrolimus ± other | 193 (23) |
| Cyclosporine + methotrexate | 576 (71) |
| Other combinations | 47 (6) |
| Use of anti-thymocyte globulin | 75 (9) |
| Transplantation year | |
| 1988–1995 | 202 (25) |
| 1995–1999 | 324 (40) |
| 2000–2008 | 290 (36) |
Data presented as no. (%) unless otherwise specified
Transplantations were facilitated through the National Marrow Donor Program (NMDP) and performed between 1988 and 2004. Data collection and analysis was performed under the auspices of the Center for International Blood and Marrow Transplant Research (CIBMTR). Pre-transplant donor and patient research samples were provided by the NMDP/CIBMTR Research Repository.
Observational studies conducted by the CIBMTR are performed in compliance with the privacy rule (HIPAA) as a Public Health Authority and in compliance with all applicable federal regulations pertaining to the protection of human research participants as determined by continuous review of the Institutional Review Boards (IRB) of the NMDP. A standardized modeling process was used, as previously described (24), to adjust for any bias introduced by the exclusion of non-consenting survivors in the NMDP cohort.
GENOTYPING
SNPs were genotyped using a previously developed in-house assay (25) based on representative SNPs for TLR1-10. SNPs where selected randomly among primarily amino acid changing SNPs, but also potentially regulatory SNPs (e.g. promotor, 3′UTR) and SNPs with previously reported functional effects from the dbSNP database (26) at the time of assay development. Twenty-nine bialleic SNPs observed in persons of European ancestry were included in the analyses (Supplemental table S1). Briefly, allele-specific primers were labelled in an allele-specific primer extension (ASPE) reaction, using polymerase chain reaction-amplified SNP-sites as their target sequences. The labelled ASPE-primers were subsequently hybridized to MicroPlex-xTAG beadsets for detection and counting on the Luminex platform (Luminex Corporation, Austin, TX, USA). All genotypings were carried out randomized and blinded to the technician performing the genotyping.
STATISTICS
Probability of leukaemia-free survival and overall survival were calculated using the Kaplan-Meier estimator. Cumulative incidences were estimated for other endpoints to accommodate competing risks. Comparison of survival curves was done using the log-rank test.
Multivariate analyses were performed using Cox proportional hazards models, which model the hazard functions for overall and leukaemia-free survivals while model the cause-specific hazards for competing risks such as TRM, relapse, aGVHD and cGVHD. All clinical variables were tested for proportional hazards assumptions using time-dependent covariate approach. Factors violating the proportional hazards assumption were adjusted through stratification. Stepwise model building procedures were performed to select the adjusted variables at 0.05 significance level for both entry and retaining in the models. Each SNP was tested for association with the clinical outcomes by forcing it into the model with the selected adjusted variables. The multivariate models are shown in Supplemental Table S2. Only 23 SNPs with minor allele frequencies ≥ 5% were included in the statistical analyses. Based on the Bonferroni criterion (23 SNPs for both patients and donors; 0.05/46) P≤0.001 was used for statistical significance in order to adjust for multiple testing. All the analyses were performed using SAS version 9.3 (The SAS Institute, Cary, NC).
RESULTS
Genotyping
The Hardy-Weinberg equilibrium (HWE) and genotype distributions were analyzed separately for patients and donors (Supplemental Table S1). All SNPs adhered to the HWE expectations at the P=0.001 level.
Six SNPs (rs3923647 in TLR1, rs5743704 in TLR2, rs5743813 in TLR6, rs5743781 in TLR7 and rs4129008 and rs466657 in TLR10) were excluded from further analyses due to minor allele frequencies < 5%. Of the remaining 23 SNPs only TLR8 rs3764879 donor type showed a significant association between genotype and outcome at the P≤0.001 level in the multivariate models (Table 2) (Supplemental table S3).
Table 2.
Association between TLR8 rs3764879 donor genotype and outcome
| Variable | Male only | Female only | ||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | n | HR (95% CI) | p | Genotype | n | HR (95% CI) | p | |
| OS | A | 378 | 1.00 | AA | 158 | 1.00 | ||
| a | 124 | 1.41 (1.09–1.83) | 0.01 | Aa | 121 | 1.13 (0.83–1.53) | 0.455 | |
| aa | 13 | 2.78 (1.43–5.41) | 0.003 | |||||
| DFS | A | 375 | 1.00 | AA | 158 | 1.00 | ||
| a | 122 | 1.45 (1.12–1.87) | 0.005 | Aa | 120 | 1.23 (0.91–1.68) | 0.188 | |
| aa | 12 | 2.34 (1.18–4.65) | 0.015 | |||||
| Relapse | A | 375 | 1.00 | AA | 158 | 1.000 | ||
| a | 122 | 1.35 (0.86–2.12) | 0.193 | Aa | 120 | 1.03 (0.55–1.91) | 0.935 | |
| aa | 12 | 0.87 (0.20–3.83) | 0.849 | |||||
| TRM | A | 145 | 1.00 | AA | 158 | 1.00 | ||
| a | 58 | 1.49 (1.09–2.04) | 0.012 | Aa | 120 | 1.21 (0.86–1.71) | 0.280 | |
| aa | 12 | 3.12 (1.44–6.74) | 0.004 | |||||
| aGVHD | ||||||||
| Grade II–IV | A | 374 | 1.00 | AA | 158 | 1.00 | ||
| a | 123 | 0.89 (0.67–1.19) | 0.427 | Aa | 118 | 1.16 (0.83–1.61) | 0.379 | |
| aa | 13 | 1.27 (0.61–2.65) | 0.529 | |||||
| Grade III–IV | A | 367 | 1.00 | AA | 148 | 1.00 | ||
| a | 121 | 0.81 (0.51–1.27) | 0.357 | Aa | 114 | 1.76 (1.04–2.98) | 0.036 | |
| aa | 13 | 3.96 (1.57–9.96) | 0.004 | |||||
| cGVHD | A | 371 | 1.00 | AA | 156 | 1.00 | ||
| a | 123 | 1.05 (0.79–1.41) | 0.718 | Aa | 120 | 0.99 (0.72–1.36) | 0.965 | |
| aa | 13 | 1.22 (0.43–3.50) | 0.709 | |||||
OS, overall survival; DFS, disease free survival; TRM, treatment related mortality; aGVHD, acute graft versus host disease; cGVHD, chronic graft versus host disease; A, Major allele hemizygous; a, minor allele hemizygous; AA, major allele homozygous; Aa, heterozygous; aa, minor allele homozygous; HR, hazard ratio; CI, confidence interval; NA, not available.
Association between donor TLR8 rs3764879 genotype and outcome
Eight-hundred-two donors were successfully genotyped for rs3764879 which is located on chromosome X. The minor allele frequency was 25% both among male and female donors. As TLR8 is located on the X chromosome male donors can only be hemizygous for a TLR8 gene, and therefore only present 2 genotypes, namely presence or absence of the major allele. Of 508 male donors 383 (75%) were hemizygous for the rs3764879 major allele while 125 (25%) were hemizygous for the minor allele. Of 294 female donors 159 (54%), 122 (42%) and 13 (4%) were homozygous for the major allele, heterozygous and homozygous for the minor allele, respectively.
In the multivariate analyses male donor hemizygosity and female donor homozygosity for the rs3764879 minor allele was an independent risk factor significantly associated with DFS (hazard ratio (HR) 1.47 (95% confidence interval (CI) 1.16–1.85); P=0.001), ++++which translated into trends towards lower OS (HR 1.44 (95% CI 1.14–1.82), P=0.002) and increased TRM (HR 1.59 (95% CI 1.19–2.12), P=0.002) (Supplemental table S3). There were no significant associations between rs3764879 genotype and relapse or GVHD (Supplemental table S3).
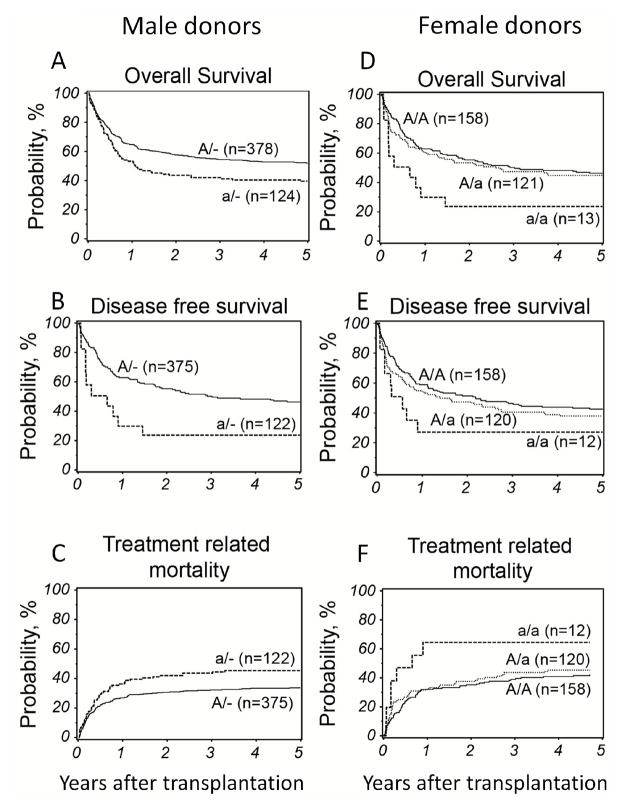
Due to the location of TLR8 on the X chromosome the multivariate analyses of the whole cohort were confounded by the minor allele disparity conferred by gender. When male and female donor rs3764879 genotypes were analyzed separately results were similar to the whole cohort (Table 2). Although not significant at the Bonferroni corrected P≤0.001 level, both male donor hemizygosity and female donor homozygosity for the minor allele were associated with OS (male donor: HR 1.41 (95% CI 1.09–1.83), P=0.010); female donor: (HR 2.78 (95% CI 1.43–5.41), P=0.003), DFS (male donor: HR 1.45 (95% CI 1.12–1.87), P=0.005; female donor: HR 2.34 (95% CI 1.18–4.65), P=0.015) and TRM (male donor: HR 1.49 (95% CI 1.09–2.04), P=0.012; female donor: HR 3.12 (95% CI 1.44–6.74), P=0.004), (Table 2 and Figure 1). Causes of death are shown in detail in table 3. Thirty-four percent of patients transplanted with a male donor hemizygous for the minor allele died of causes unrelated to relapse or GVHD, namely interstitial pneumonia, infection or organ failure, while only 27% of patients transplanted with a donor hemizygous for the major allele died of these causes (p=0.64). In the group of patients transplanted with a female donor homozygous for the minor allele, heterozygous or homozygous for the major allele, 39%, 33% and 28% (p=0.68), respectively, died of interstitial pneumonia, infection or organ failure.
Figure 1.
Adjusted probability of overall survival, disease free survival and treatment related mortality according donor TLR8 rs3764879 genotype analyzed separately for male (panel A, B and C) and female donors (panel D, E and F). A/−, Major allele hemizygous (solid line); a/−, minor allele hemizygous (dashed line); A/A, major allele homozygous (solid line); A/a, heterozygous (dotted line); a/a, minor allele homozygous (dashed line).
Table 3.
Causes of death according to TLR8 rs3764879 donor genotype
| Cause of death | Male donors | Female donors | ||||
|---|---|---|---|---|---|---|
| Genotype | N (%) | p | Genotype | N (%) | p | |
| Primary disease | A/- | 40 (10) | 0.76 | A/A | 17 (11) | 0.37 |
| a/- | 17 (14) | A/a | 8 (7) | |||
| a/a | 1 (8) | |||||
| New malignancy | A/- | 5 (1) | 0.53 | A/A | 1 (1) | 0.92 |
| a/- | 1 (1) | A/a | 1 (1) | |||
| a/a | 0 (0) | |||||
| Graft versus host disease | A/- | 29 (8) | 0.91 | A/A | 17 (11) | 0.72 |
| a/- | 11 (9) | A/a | 10 (8) | |||
| a/a | 2 (15) | |||||
| Interstitial pneumonia | A/- | 21 (6) | 0.41 | A/A | 16 (16) | 0.74 |
| a/- | 11 (9) | A/a | 16 (13) | |||
| a/a | 2 (15) | |||||
| Infection | A/- | 45 (12) | 0.77 | A/A | 20 (13) | 0.44 |
| a/- | 19 (15) | A/a | 10 (8) | |||
| a/a | 2 (15) | |||||
| Organ failure | A/- | 37 (10) | 0.70 | A/A | 9 (6) | 0.19 |
| a/- | 13 (10) | A/a | 14 (12) | |||
| a/a | 1 (8) | |||||
| Other | A/- | 29 (8) | 0.50 | A/A | 16 (10) | 0.58 |
| a/- | 9 (7) | A/a | 16 (13) | |||
| a/a | 3 (23) | |||||
N, number of deaths; percentage, percent of all deaths; A, Major allele hemizygous; a, minor allele hemizygous; AA, major allele homozygous; Aa, heterozygous; aa, minor allele homozygous.
No associations between genotype and relapse or GVHD were observed. The complete multivariate analyses including all covariates are shown in Supplemental table S4). Of note the data from female donors should be interpreted cautiously as the number of female donors homozygous for the minor allele was low (Table 2).
TLR1, 4, 6 and 9 SNPs previously associated with outcome in allogeneic HCT
Heterozygosity for the TLR4 SNP rs4986790 has previously been associated with hemorrhagic cystitis and invasive aspergillosis (23;27;28). Due to low numbers of homozygous minor allele carriers both in the patient (n=1) and donor (n=3) cohorts these were included in the group of heterozygotes for analysis. Patient minor allele carriage of the TLR4 rs4986790 SNP tended to be associated with increased risk of TRM (HR 1.15 (95% CI 1.12–2.04); P=0.007) translating into lower DFS (HR 1.36 (95% CI 1.05–1.77); P=0.0214) and OS (HR 1.42 (95% CI 1.09–1.86); P=0.010) (Figure 2 and Table 4). There were no associations between the rs4986790 genotype and relapse or GVHD.
Table 4.
Association between TLR1, 4, 6 and 9 genotype and outcome after allogeneic HCT
| Variable | Genotype | Patient TLR1 (rs5743611) | Patient TLR4 (rs4986790) | ||||
|---|---|---|---|---|---|---|---|
| n | HR (95% CI) | p | n | HR (95% CI) | p | ||
| OS | AA | 666 | 1.000 | 696 | 1.000 | ||
| Aa | 114 | 1.017 (0.779–1.328) | 0.902 | 96 | 1.424 (1.089–1.863) | 0.010 | |
| aa | 12 | 1.432 (0.729–2.813 | 0.297 | 0 | NA | NA | |
| DFS | AA | 662 | 1.000 | 689 | 1.000 | ||
| Aa | 112 | 0.988 (0.758–1.286) | 0.927 | 96 | 1.363 (1.047–1.774) | 0.0214 | |
| aa | 11 | 1.195 (0.585–2.443) | 0.625 | 0 | NA | NA | |
| Relapse | AA | 662 | 1.000 | 689 | 1.000 | ||
| Aa | 112 | 0.945 (0.584–1.530) | 0.819 | 96 | 1.034 (0.592–1.805) | 0.907 | |
| aa | 11 | 2.054 (0.740–5.704) | 0.167 | 0 | NA | NA | |
| TRM | AA | 662 | 1.000 | 689 | 1.000 | ||
| Aa | 112 | 0.950 (0.692–1.304) | 0.751 | 96 | 1.515 (1.123.2.042) | 0.007 | |
| aa | 11 | 0.860 (0.316–2.335) | 0.767 | 0 | NA | NA | |
| aGVHD | |||||||
| Grade II–IV | AA | 659 | 1.000 | 689 | 1.000 | ||
| Aa | 114 | 1.231 (0.947–0.294) | 0.121 | 96 | 1.120 (0.839–1.495) | 0.442 | |
| aa | 12 | 0.714 (0.294–1.735) | 0.457 | 0 | NA | NA | |
| Grade III–IV | AA | 638 | 1.000 | 668 | 1.000 | ||
| Aa | 112 | 1.553 (1.054–2.290) | 0.026 | 94 | 1.071 (0.686–1.672) | 0.763 | |
| aa | 12 | 0.267 (0.037–1.935) | 0.191 | 0 | NA | NA | |
| cGVHD | AA | 650 | 1.000 | 682 | 1.000 | ||
| Aa | 112 | 1.064 (0.807–1.403) | 0.660 | 92 | 0.789 (0.561–1.109) | 0.172 | |
| aa | 12 | 1.026 (0.451–2.336) | 0.951 | 0 | NA | NA | |
For TLR1 SNP rs5743611 which has previously been associated with invasive aspergillosis, our data were suggestive of an association between patient heterozygosity and grade III–IV acute GVHD was observed (HR 1.55 (95% CI 1.05–2.29); P=0.026), with no impact on survival (Table 4).
In contrast to previous reports no significant associations between the TLR6 SNP rs5743810 (29) or the TLR9 SNP rs187084 (30) and outcome were observed (Table 4).
Of note, none of the previously investigated SNPs described in this section were significantly associated with outcome in the current study as the level for statistical significance was set at the conservative Bonferroni corrected P≤0.001.
DISCUSSION
The current study is the largest investigation of associations between TLR SNPs and outcome after allogeneic HCT at present. Our cohort of 816 patient and donor pairs was genotyped for 29 SNPs across the 10 known human TLR genes. To achieve sufficient statistical power SNPs with minor allele frequencies <5% were excluded, leaving 23 SNPs to be tested in the multivariate models for clinical outcome variables. Although conservative, the Bonferroni method was used to adjust for multiple comparisons to lower the risk of false positive associations. As TLR8 is located on the X chromosome and males only carry one of these they can only present phenotypes corresponding to either major or minor allele homozygosity, while heterozygosity also exists in the female population. Hemi-or homozygosity for the rs3764879 minor allele in male and female donors was significantly associated with lower DFS with trends towards lower OS and TRM. No association with GVHD or relapse was observed. Although the rs3764879 genotype is inherently confounded by gender no interactions between gender and genotype were observed. When male and female donors were analyzed separately, patterns similar to that seen in the whole cohort were observed, but levels of significance were lower in the separate male and female donor subsets, in line with the reduced power of the stratified analyses. Especially in the female subset, data should be interpreted with caution due to low numbers of minor allele homozygous donors. However the data do suggest a clinical impact of to the absence of phenotypical expression of the rs3764879 major allele.
Althougn non significant, we observed increased frequencies of death due interstitial pneumonia, infection or organ failure in patients transplanted with minor allele hemi-or homozygous donors, which is iin agreement
No associations were observed between rs3764879 and relapse or GVHD, indicating that the poorer outcome was conferred by other causes. Although not significant, we did observe higher frequencies of death related to interstitial pneumonia, infection or organ failure. The finding is in line with TLR8 being part of the anti-viral immune response where it recognizes non-self nucleic acids and subsequently stimulates the release of pro-inflammatory cytokines (31). The rs3764879 SNP is located in the promoter region of TLR8 and the minor allele has been associated with lower cytokine production (32). TLR8 is expressed mainly by haematopoietically derived cells (monocytes and myeloid derived dendritic cells) (33;34) and in keeping with this, the significant associations between outcomes and genotypes were not found for recipient genotypes of the TLR8 SNP.
rs3764879 is in complete linkage disequilibrium with the codon 1 TLR8 SNP rs3764880 SNP, which introduces a frame-shift mutation leading to a truncated final protein (35;36). Only few studies have adressed the association between TLR8 genotype and disease, and in agreement with our data they have shown associations between the minor allele of the promoter SNP (rs3764879) or the codon 1 SNP (rs3764880) and progression of HIV infection, susceptibility to mycobacterium turberculosis, hepatitis C infection and asthma (36–40).
Several polymorphisms across most TLRs have been studied in a variety of disease settings (41). The most extensively investigated are the functional non-synonymous TLR4 SNPs D299G (rs4986790) and T399I (rs4986791), which are in linkage disequilibrium (21). TLR4 recognizes microbial cell wall components from gram negative bacteria and fungal ligands such as candida mannan, glucuronoxylomannan and Aspergillus fumigatus antigens (9;42;43). In the setting of allogeneic HCT both D299G and T399I have been associated with invasive aspergillosis. Koldenhoff et al. observed that patient or donor carriage of the minor allele of either SNP was associated with invasive aspergillosis (27), while Bochud et al. in a large discovery/validation cohort observed a significant association between donor carriage of the minor allele and invasive aspergillosis and TRM (21). In contrast Kesh et al. did not observe an association between TLR4 D299G and T399I genotype and invasive aspergillosis. The same study also included SNPs in TLR1 and TLR6 which heterodimerize with TLR2 to mediate responses to lipopeptides from several pathogens. Presence of either the minor allele of TLR1(rs5743611) or TLR1 (rs4833095) and TLR6 (rs5743810) in the recipient were associated with increased risk of invasive aspergillosis (29).
Presence of the minor allele of TLR4 D299G in patient and donor has also been associated with hemorrhagic cystitis in a pediatric cohort transplanted after myeloablative conditioning (23).
Elmaagcli et al. investigated 2 SNPs in TLR9, which is located intracellularly and senses single stranded DNA from microbial pathogens containing CpG motifs, in a cohort of AML patients treated with high dose allogeneic HCT (30). They observed that patient homozygosity for rs187084 conferred a lower 5 year probability of relapse and increased OS.
None of the previously published associations between TLR genotypes and allogeneic HCT were replicated at the Bonferroni adjusted significance level of P≤0.001. However in the current study, patient carriage of the TLR4 SNP rs4986790 minor allele was associated with TRM and OS at the P<0.05 level, lending support to the probable importance of TLR4 genotype on infection related morbidity and mortality. In contrast to previous reports no association between D299G and acute GVHD were observed (20;28), and no relevant significant associations between previously studied SNPs in TLR1, 6 and 9 were observed (29;30).
The inconsistent results often observed across genetic association studies are due to several factors that make direct interstudy comparisons difficult. The generally small cohorts in transplant studies increase the risk of type I errors as the effect of single genetic variants usually is modest. Furthermore heterogeneity between patient populations, with differences in treatment regimens, diagnoses, racial admixture, other yet unknown risk factors and the possibility of the selected polymorphisms being in linkage disequilibrium with unknown functional polymorphisms all contribute to an unclear picture.
In conclusion the present study is currently the largest and most comprehensive investigation of associations between TLR genotype and outcome after allogeneic HCT. Although none of the previously published associations between TLR SNPs and outcome were validated at the Bonferroni corrected significance levels, similar trends were observed. However a novel association between a TLR8 promoter polymorphism and survival was observed, and evidence supporting the importance of TLR4 D299G was presented. To confirm the significance of these findings further experimental and clinical studies are needed to explain their molecular background and assess their impact on outcome in a prospective manner, respectively.
Supplementary Material
Supplemental Table S1. Genotype distribution and HWE of included SNPs.
Supplemental Table S2. Multivariate models based on clinical variables.
Supplemental Table S3. Multivariate Cox regression analyzes of single SNPs and outcome
Supplemental Table S4. Multivariate Cox regression analyzes of TLR8 rs37648879 genotype and outcome for male and females donors separately
Table 5.
| Variable | Donor TLR4 (rs4986790) | Patient TLR6 (rs5743810) | Patient TLR9 (rs187084) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | HR (95% CI) | p | n | HR (95% CI) | p | n | HR (95% CI) | p | ||
| OS | 703 | 1.000 | 247 | 1.000 | 278 | 1.000 | ||||
| 102 | 1.204 (0.928–1.562) | 0.163 | 409 | 1.073 (0.871–1.321) | 0.510 | 386 | 0.877 (0.716–1.073) | 0.202 | ||
| 0 | NA | NA | 136 | 1.065 (0.810–1.401 | 0.652 | 128 | 0.920 (0.700–1.208) | 0.549 | ||
| DFS | 697 | 1.000 | 245 | 1.000 | 274 | 1.000 | ||||
| 101 | 1.205 (0.935–1.554) | 0.150 | 404 | 1.039 (0.846–1.275) | 0.716 | 385 | 0.865 (0.709–1.056) | 0.153 | ||
| 0 | NA | NA | 136 | 1.052 (0.804–1.376) | 0.711 | 126 | 0.927 (0.709–1.211) | 0.580 | ||
| Relapse | 697 | 1.000 | 245 | 1.000 | 274 | 1.000 | ||||
| 101 | 0.734 (0.428–1.260) | 0.262 | 404 | 0.705 (0.484–1.028) | 0.069 | 385 | 0.761 (0.531–1.093) | 0.139 | ||
| 0 | NA | NA | 136 | 0.978 (0.614–1.557) | 0.925 | 126 | 0.622 (0.360–1.073) | 0.088 | ||
| TRM | 697 | 1.000 | 245 | 1.000 | 274 | 1.000 | ||||
| 101 | 1.388 (1.040–1.854) | 0.026 | 404 | 1.188 (0.929–1.518) | 0.169 | 385 | 0.961 (0.758–1.220) | 0.746 | ||
| 0 | NA | NA | 136 | 1.032 (0.744–1.431) | 0.851 | 126 | 1.061 (0.779–1.444) | 0.709 | ||
| aGVHD | 698 | 1.000 | 244 | 1.000 | 275 | 1.000 | ||||
| Grade II–IV | 99 | 1.018 (0.764–1.358) | 0.901 | 407 | 0.988 (0.797–1.224) | 0.912 | 283 | 0.982 (0.792–1.217) | 0.865 | |
| 0 | NA | NA | 134 | 0.812( 0.603–1.092) | 0.168 | 127 | (0.889–1.555) | 0.258 | ||
| 676 | 1.000 | 239 | 1.000 | 270 | 1.000 | |||||
| Grade III–IV | 98 | 1.286 (0.849–1.946) | 0.235 | 395 | 0.862 (0.620–1.198) | 369 | 0.809 (0.579–1.128) | 0.212 | ||
| 0 | NA | NA | 128 | 0.809 (0.512–1.278) | 123 | 1.071 (0.712–1.613) | 0.741 | |||
| 687 | 1.000 | 244 | 1.000 | 271 | 1.000 | |||||
| cGVHD | 99 | 1.089 (0.806–1.470) | 0.580 | 395 | 0.973 (0.785–1.207) | 0.805 | 378 | 1.128 (0.909–1.400) | 0.275 | |
| 0 | NA | NA | 135 | 0.791 (0.586–1.067) | 0.125 | 125 | 1.193 (0.898–1.585) | 0.223 | ||
OS, overall survival; DFS, disease free survival; TRM, treatment related mortality; aGVHD, acute graft versus host disease; cGVHD, chronic graft versus host disease; AA, major allele homozygous; Aa, heterozygous; aa, minor allele homozygous; HR, hazard ratio; CI, confidence interval; Rs number, reference single nucleotide polymorphism number; NA, not available.
Acknowledgments
The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U10HL069294 from NHLBI and NCI; a contract HHSH250201200016C with Health Resources and Services Administration (HRSA/DHHS); two Grants N00014-13-1-0039 and N00014-14-1-0028 from the Office of Naval Research; and grants from Actinium Pharmaceuticals; Allos Therapeutics, Inc.; Amgen, Inc.; Anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Celgene Corporation; Chimerix, Inc.; Fred Hutchinson Cancer Research Center; Fresenius-Biotech North America, Inc.; Gamida Cell Teva Joint Venture Ltd.; Genentech, Inc.; Gentium SpA; Genzyme Corporation; GlaxoSmithKline; Health Research, Inc. Roswell Park Cancer Institute; HistoGenetics, Inc.; Incyte Corporation; Jeff Gordon Children’s Foundation; Kiadis Pharma; The Leukemia & Lymphoma Society; Medac GmbH; The Medical College of Wisconsin; Merck & Co, Inc.; Millennium: The Takeda Oncology Co.; Milliman USA, Inc.; Miltenyi Biotec, Inc.; National Marrow Donor Program; Onyx Pharmaceuticals; Optum Healthcare Solutions, Inc.; Osiris Therapeutics, Inc.; Otsuka America Pharmaceutical, Inc.; Perkin Elmer, Inc.; Remedy Informatics; Sanofi US; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix, Inc.; St. Baldrick’s Foundation; StemCyte, A Global Cord Blood Therapeutics Co.; Stemsoft Software, Inc.; Swedish Orphan Biovitrum; Tarix Pharmaceuticals; TerumoBCT; Teva Neuroscience, Inc.; THERAKOS, Inc.; University of Minnesota; University of Utah; and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
The study was also supported by grants from the Danish Biotechnology Program, the Danish Child Cancer Foundation and the Dagmar Marshall Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
BK analyzed and interpreted data and drafted and revised the manuscript and. CE genotyped samples, analyzed and interpreted data and revised the manuscript. SS, MH, TW and SJL provided patient and donor material, performed statistical analyses, interpreted data and revised the manuscript. KM planned and designed the study, interpreted data and revised the manuscript.
FINANCIAL DISCLOSURES
The authors report no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009 May 2;373(9674):1550–61. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleakley M, Riddell SR. Molecules and mechanisms of the graft-versus-leukaemia effect. Nat Rev Cancer. 2004 May;4(5):371–80. doi: 10.1038/nrc1365. [DOI] [PubMed] [Google Scholar]
- 3.Petersdorf EW, Gooley TA, Anasetti C, Martin PJ, Smith AG, Mickelson EM, et al. Optimizing outcome after unrelated marrow transplantation by comprehensive matching of HLA class I and II alleles in the donor and recipient. Blood. 1998 Nov 15;92(10):3515–20. [PubMed] [Google Scholar]
- 4.Lin MT, Storer B, Martin PJ, Tseng LH, Grogan B, Chen PJ, et al. Genetic variation in the IL-10 pathway modulates severity of acute graft-versus-host disease following hematopoietic cell transplantation: synergism between IL-10 genotype of patient and IL-10 receptor beta genotype of donor. Blood. 2005 Dec 1;106(12):3995–4001. doi: 10.1182/blood-2004-11-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamim Z, Ryder LP, Christensen IJ, Toubert A, Norden J, Collin M, et al. Prognostic significance of interleukin-7 receptor-alpha gene polymorphisms in allogeneic stem-cell transplantation: a confirmatory study. Transplantation. 2011 Apr 15;91(7):731–6. doi: 10.1097/TP.0b013e31820f08b2. [DOI] [PubMed] [Google Scholar]
- 6.Holler E, Rogler G, Herfarth H, Brenmoehl J, Wild PJ, Hahn J, et al. Both donor and recipient NOD2/CARD15 mutations associate with transplant-related mortality and GvHD following allogeneic stem cell transplantation. Blood. 2004 Aug 1;104(3):889–94. doi: 10.1182/blood-2003-10-3543. [DOI] [PubMed] [Google Scholar]
- 7.Mullighan CG, Heatley SL, Danner S, Dean MM, Doherty K, Hahn U, et al. Mannose-binding lectin status is associated with risk of major infection following myeloablative sibling allogeneic hematopoietic stem cell transplantation. Blood. 2008 Sep 1;112(5):2120–8. doi: 10.1182/blood-2007-07-100222. [DOI] [PubMed] [Google Scholar]
- 8.Kornblit B, Masmas T, Petersen SL, Madsen HO, Heilmann C, Schejbel L, et al. Association of HMGB1 polymorphisms with outcome after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2010 Feb;16(2):239–52. doi: 10.1016/j.bbmt.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006 Feb 24;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J Biol Chem. 2004 Feb 27;279(9):7370–7. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 11.Kokkola R, Andersson A, Mullins G, Ostberg T, Treutiger CJ, Arnold B, et al. RAGE is the major receptor for the proinflammatory activity of HMGB1 in rodent macrophages. Scand J Immunol. 2005 Jan;61(1):1–9. doi: 10.1111/j.0300-9475.2005.01534.x. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov S, Dragoi AM, Wang X, Dallacosta C, Louten J, Musco G, et al. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007 Sep 15;110(6):1970–81. doi: 10.1182/blood-2006-09-044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu SF, Malik AB. NF-kappa B activation as a pathological mechanism of septic shock and inflammation. Am J Physiol Lung Cell Mol Physiol. 2006 Apr;290(4):L622–L645. doi: 10.1152/ajplung.00477.2005. [DOI] [PubMed] [Google Scholar]
- 14.Xun CQ, Thompson JS, Jennings CD, Brown SA, Widmer MB. Effect of total body irradiation, busulfan-cyclophosphamide, or cyclophosphamide conditioning on inflammatory cytokine release and development of acute and chronic graft-versus-host disease in H-2-incompatible transplanted SCID mice. Blood. 1994 Apr 15;83(8):2360–7. [PubMed] [Google Scholar]
- 15.Hill GR, Crawford JM, Cooke KR, Brinson YS, Pan L, Ferrara JL. Total body irradiation and acute graftversus-host disease: the role of gastrointestinal damage and inflammatory cytokines. Blood. 1997 Oct 15;90(8):3204–13. [PubMed] [Google Scholar]
- 16.D’Agostini C, Pica F, Febbraro G, Grelli S, Chiavaroli C, Garaci E. Antitumour effect of OM-174 and cyclophosphamide on murine B16 melanoma in different experimental conditions. Int Immunopharmacol. 2005 Jul;5(7–8):1205–12. doi: 10.1016/j.intimp.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Spaner DE, Shi Y, White D, Shaha S, He L, Masellis A, et al. A phase I/II trial of TLR-7 agonist immunotherapy in chronic lymphocytic leukemia. Leukemia. 2010 Jan;24(1):222–6. doi: 10.1038/leu.2009.195. [DOI] [PubMed] [Google Scholar]
- 18.Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008 Jan 7;27(2):161–7. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 19.Dorn A, Kippenberger S. Clinical application of CpG-, non-CpG-, and antisense oligodeoxynucleotides as immunomodulators. Curr Opin Mol Ther. 2008 Feb;10(1):10–20. [PubMed] [Google Scholar]
- 20.Elmaagacli AH, Koldehoff M, Hindahl H, Steckel NK, Trenschel R, Peceny R, et al. Mutations in innate immune system NOD2/CARD 15 and TLR-4 (Thr399Ile) genes influence the risk for severe acute graft-versus-host disease in patients who underwent an allogeneic transplantation. Transplantation. 2006 Jan 27;81(2):247–54. doi: 10.1097/01.tp.0000188671.94646.16. [DOI] [PubMed] [Google Scholar]
- 21.Bochud PY, Chien JW, Marr KA, Leisenring WM, Upton A, Janer M, et al. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med. 2008 Oct 23;359(17):1766–77. doi: 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sivula J, Cordova ZM, Tuimala J, Jaatinen T, Partanen J, Volin L, et al. Toll-like receptor gene polymorphisms confer susceptibility to graft-versus-host disease in allogenic hematopoietic stem cell transplantation. Scand J Immunol. 2012 Sep;76(3):336–41. doi: 10.1111/j.1365-3083.2012.02737.x. [DOI] [PubMed] [Google Scholar]
- 23.Gruhn B, Kloppner N, Pfaffendorf-Regler N, Beck J, Zintl F, Bartholoma S, et al. Toll-like 4 receptor variant, Asp299Gly, and reduced risk of hemorrhagic cystitis after hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2012 Jun;18(6):958–63. doi: 10.1016/j.bbmt.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Farag SS, Bacigalupo A, Eapen M, Hurley C, Dupont B, Caligiuri MA, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006 Aug;12(8):876–84. doi: 10.1016/j.bbmt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Enevold C, Oturai AB, Sorensen PS, Ryder LP, Koch-Henriksen N, Bendtzen K. Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. J Neuroimmunol. 2009 Jul 25;212(1–2):125–31. doi: 10.1016/j.jneuroim.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001 Jan 1;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koldehoff M, Beelen DW, Elmaagacli AH. Increased susceptibility for aspergillosis and posttransplant immune deficiency in patients with gene variants of TLR4 after stem cell transplantation. Transpl Infect Dis. 2013 Oct;15(5):533–9. doi: 10.1111/tid.12115. [DOI] [PubMed] [Google Scholar]
- 28.Lorenz E, Schwartz DA, Martin PJ, Gooley T, Lin MT, Chien JW, et al. Association of TLR4 mutations and the risk for acute GVHD after HLA-matched-sibling hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2001;7(7):384–7. doi: 10.1053/bbmt.2001.v7.pm11529488. [DOI] [PubMed] [Google Scholar]
- 29.Kesh S, Mensah NY, Peterlongo P, Jaffe D, Hsu K, VAN DEN BM, et al. TLR1 and TLR6 polymorphisms are associated with susceptibility to invasive aspergillosis after allogeneic stem cell transplantation. Ann N Y Acad Sci. 2005 Dec;1062:95–103. doi: 10.1196/annals.1358.012. [DOI] [PubMed] [Google Scholar]
- 30.Elmaagacli AH, Steckel N, Ditschkowski M, Hegerfeldt Y, Ottinger H, Trenschel R, et al. Toll-like receptor 9, NOD2 and IL23R gene polymorphisms influenced outcome in AML patients transplanted from HLA-identical sibling donors. Bone Marrow Transplant. 2011 May;46(5):702–8. doi: 10.1038/bmt.2010.166. [DOI] [PubMed] [Google Scholar]
- 31.Cervantes JL, Weinerman B, Basole C, Salazar JC. TLR8: the forgotten relative revindicated. Cell Mol Immunol. 2012 Nov;9(6):434–8. doi: 10.1038/cmi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang CH, Eng HL, Lin KH, Liu HC, Chang CH, Lin TM. Functional polymorphisms of TLR8 are associated with hepatitis C virus infection. Immunology. 2014 Apr;141(4):540–8. doi: 10.1111/imm.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000 Sep;11(3):372–8. [PubMed] [Google Scholar]
- 34.Ito T, Wang YH, Liu YJ. Plasmacytoid dendritic cell precursors/type I interferon-producing cells sense viral infection by Toll-like receptor (TLR) 7 and TLR9. Springer Semin Immunopathol. 2005 Jan;26(3):221–9. doi: 10.1007/s00281-004-0180-4. [DOI] [PubMed] [Google Scholar]
- 35.Cheng PL, Eng HL, Chou MH, You HL, Lin TM. Genetic polymorphisms of viral infection-associated Toll-like receptors in Chinese population. Transl Res. 2007 Nov;150(5):311–8. doi: 10.1016/j.trsl.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Oh DY, Taube S, Hamouda O, Kucherer C, Poggensee G, Jessen H, et al. A functional toll-like receptor 8 variant is associated with HIV disease restriction. J Infect Dis. 2008 Sep 1;198(5):701–9. doi: 10.1086/590431. [DOI] [PubMed] [Google Scholar]
- 37.Davila S, Hibberd ML, Hari DR, Wong HE, Sahiratmadja E, Bonnard C, et al. Genetic association and expression studies indicate a role of toll-like receptor 8 in pulmonary tuberculosis. PLoS Genet. 2008 Oct;4(10):e1000218. doi: 10.1371/journal.pgen.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, Borglum AD. Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax. 2008 Dec;63(12):1064–9. doi: 10.1136/thx.2007.094128. [DOI] [PubMed] [Google Scholar]
- 39.Wang CH, Eng HL, Lin KH, Chang CH, Hsieh CA, Lin YL, et al. TLR7 and TLR8 gene variations and susceptibility to hepatitis C virus infection. PLoS One. 2011;6(10):e26235. doi: 10.1371/journal.pone.0026235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalgic N, Tekin D, Kayaalti Z, Cakir E, Soylemezoglu T, Sancar M. Relationship between toll-like receptor 8 gene polymorphisms and pediatric pulmonary tuberculosis. Dis Markers. 2011;31(1):33–8. doi: 10.3233/DMA-2011-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misch EA, Hawn TR. Toll-like receptor polymorphisms and susceptibility to human disease. Clin Sci (Lond) 2008 Mar;114(5):347–60. doi: 10.1042/CS20070214. [DOI] [PubMed] [Google Scholar]
- 42.Braedel S, Radsak M, Einsele H, Latge JP, Michan A, Loeffler J, et al. Aspergillus fumigatus antigens activate innate immune cells via toll-like receptors 2 and 4. Br J Haematol. 2004 May;125(3):392–9. doi: 10.1111/j.1365-2141.2004.04922.x. [DOI] [PubMed] [Google Scholar]
- 43.Shoham S, Huang C, Chen JM, Golenbock DT, Levitz SM. Toll-like receptor 4 mediates intracellular signaling without TNF-alpha release in response to Cryptococcus neoformans polysaccharide capsule. J Immunol. 2001 Apr 1;166(7):4620–6. doi: 10.4049/jimmunol.166.7.4620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Genotype distribution and HWE of included SNPs.
Supplemental Table S2. Multivariate models based on clinical variables.
Supplemental Table S3. Multivariate Cox regression analyzes of single SNPs and outcome
Supplemental Table S4. Multivariate Cox regression analyzes of TLR8 rs37648879 genotype and outcome for male and females donors separately