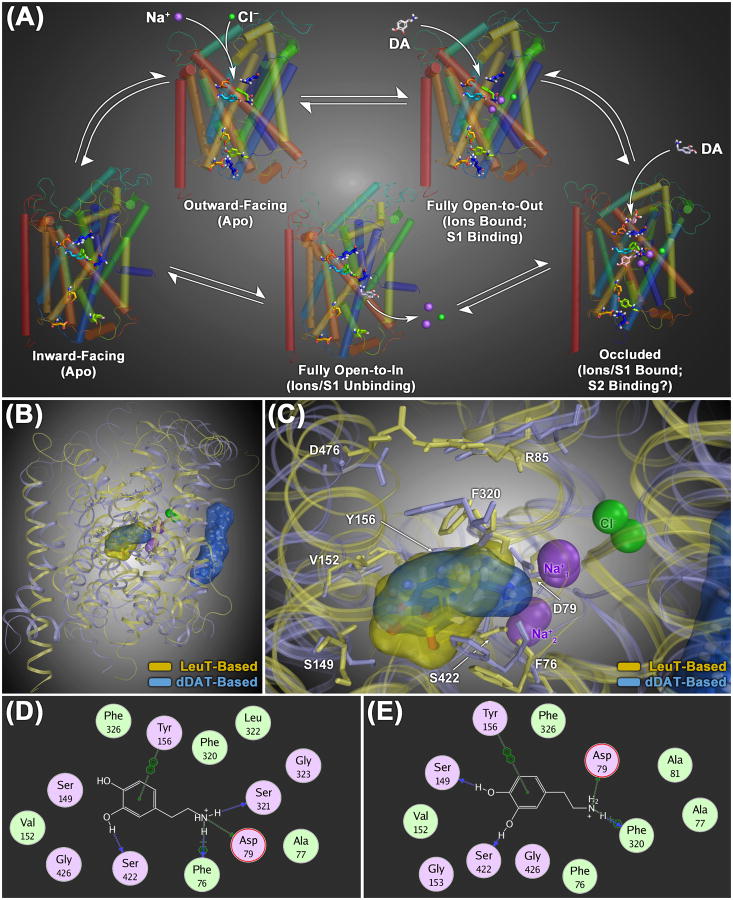
Figure 1.
(A) Model of the conformational cycle for substrate translocation by the dopamine transporter (DAT), based upon crystal structures of the bacterial NSS family protein LeuT. In its default ligand-free (apo) configuration, the transporter protein is thought to be in dynamic equilibrium between outward- and inward-facing conformational states (upper left and lower left structures, respectively). Binding of extracellular Na+ ions at the S1 site stabilizes an open-to-out conformation with a fully open extracellular gate (upper right structure), allowing substrate molecules maximum access to the core S1 binding domain. Substrate binding at the S1 site induces closure of the extracellular gate, establishing an occluded, closed-to-out conformation (lower right structure). It has been suggested that interaction of a second substrate molecule with a secondary binding domain in the extracellular vestibule (the S2 site, located 11-13 Å above the S1 site) helps facilitate opening of the intracellular gating network, giving rise to a fully inward-facing (open-to-in) conformation capable of releasing the S1-bound substrate and ions into the cytosol (lower middle structure). (B) Structural overlay of homology models of the human DAT protein (hDAT), constructed using either the LeuT (yellow ribbons) or the dDAT (light blue ribbons) crystals as structural templates. Overall, the two structures show a high degree of geometric congruence. The most profound differences are the position of the second extracellular loop region (EL2) and the orientation of TM12, which is kinked in the center of the helix in the dDAT-based structure (at P572). In the dDAT-based model, a molecule of cholesterol (sticks highlighted with a translucent blue molecular surface) is shown bound to the cavity formed by TM1, TM5 and TM7, as reported for the dDAT crystal structure. (C) Zoomed-in superposition view of both the LeuT- and dDAT-based transporter models with DA docked at the S1 site (rendered as sticks with translucent yellow and blue molecular surfaces, respectively). Residues that line the S1 binding site are labeled and rendered as thin colored sticks. (D – E) Two-dimensional molecular interaction diagrams of DA bound at the S1 site of the dDAT-based model (D) and the LeuT-based model (E). For each panel, the interaction map depicts respective DAT residues located within 4.5 Å of the bound DA molecule (hydrophobic residues are colored green and polar residues are purple). The most significant (non van der Waals) DAT/ligand interactions are indicated with dotted lines and a symbol depicting the chemistry of the interaction formed: side-chain hydrogen bond (green), main-chain hydrogen bond (blue), cation-π bond (
 +) or aromatic π-stacking (
+) or aromatic π-stacking (

 ).
).