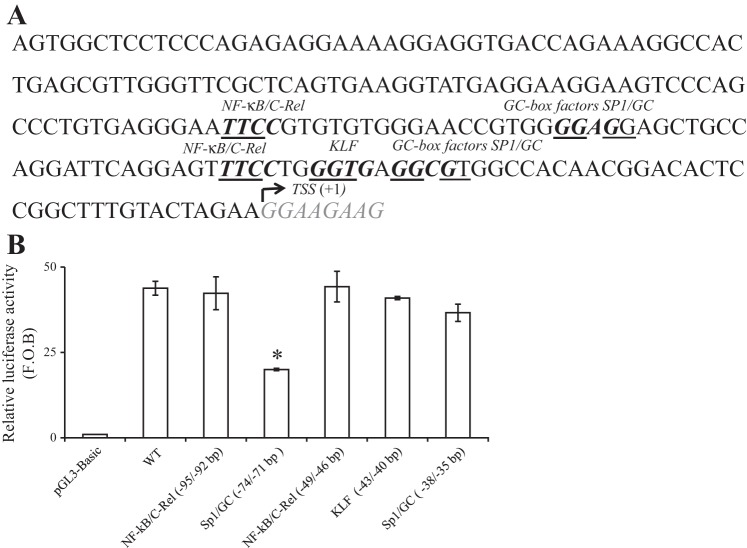
Fig. 2.
Schematic representation of the minimal promoter region of the SLC52A3 gene, and the effect of mutating putative cis-regulatory sites on promoter activity. A: nucleotide sequence of the minimal region required for basal activity of SLC52A3 is shown (38,119–37,913 on chromosome 20). Bold nucleotides indicate the positions of several identified putative cis-regulatory elements. TSS is underlined and numbered +1. B: effect of mutating specific putative sites in the SLC52A3 minimal promoter on promoter activity. Conserved nucleotides in the core consensus binding sites for stimulating protein-1 (Sp1), NF-κB/C-Rel, or Kruppel-like factor (KLF) were mutated. Mutated minimal regions were then tested for promoter activity using the luciferase assay system and HuTu 80 cells. The results are expressed as relative fold to the pGL3-basic vector and represent means ± SE of at least 3 independent experiments. *P < 0.01.