Fig. 3.
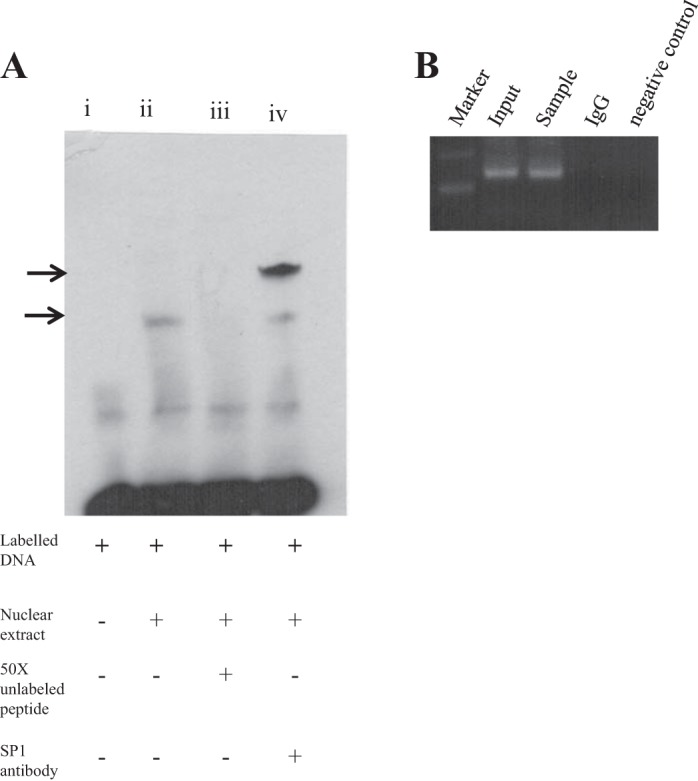
EMSA with oligonucleotide competition and chromatin immunoprecipitation (ChIP) analysis. A: gel shift assay was performed with a biotin-labeled 35-bp region of the SLC52A3 minimal promoter spanning a sequence between −89 and −55 and nuclear extract from HuTu 80 cells. A 50-fold molar excess of the same unlabeled fragment was used for oligonucleotide competition. Major DNA/protein complexes are indicated with arrows. Gel shift assay was performed by incubation with Sp1 antibody. B: binding of Sp1 transcription factor to the SLC52A3 promoter in vivo. ChIP assay was performed using the HuTu 80 cells and the antibodies against Sp1. The immunoprecipitated DNA was PCR amplified using primers specific for the region containing Sp1. A control reaction with normal IgG was used as a negative control.
