Figure 6.
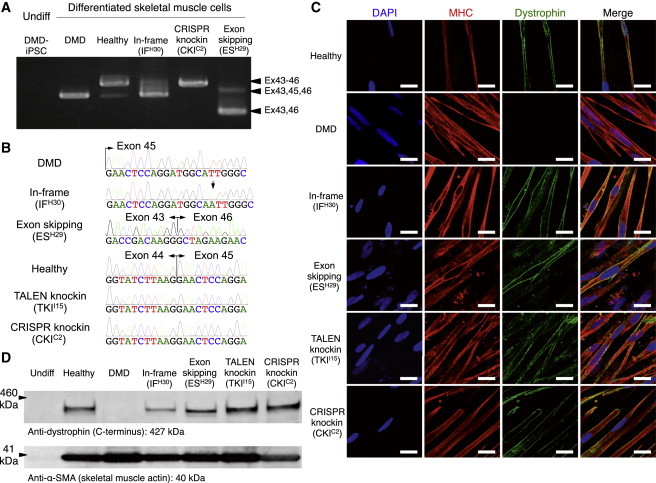
Restoration of the Dystrophin Protein in Differentiated Myogenic Cells
(A) RT-PCR analysis for dystrophin cDNA from iPSCs and cells differentiated from the corrected clones toward skeletal muscle lineage. The original DMD patient and an IF clone (IFH30) with 1 bp insertion corresponded to the 452 and 453 bp PCR bands, respectively. The healthy control and knockin clones (TALEN-mediated TKII15 and CRISPR-mediated CKIC2) corresponded to the 600 bp bands; and the ES clone (ESH29) corresponded to the 276 bp band.
(B) Sanger sequence analysis of dystrophin cDNAs from differentiated skeletal muscle cells. The IF clone (IFH30) exhibited a 1 bp insertion (A, black arrow), the ES clone (ESH29) exhibited the conjugation of exons 43 and 46 due to the skipping of exon 45, and knockin clones (TKII15 and CKIC2) exhibited the complete restoration of exon 44 in front of exon 45 as the healthy control.
(C) Immunofluorescence staining of skeletal muscle cells differentiated from the corrected clones. A z axis section of the confocal microscopy image shows submembrane localization of the dystrophin protein in the healthy control and all corrected clones, but not in the uncorrected original DMD iPSCs. The cells were stained by DAPI, a marker of skeletal differentiation (myosin heavy chain [MHC]), in red and an antibody that detects the rod domain of dystrophin (DYS1) in green. Scale bar, 50 μm.
(D) Western blot analysis to estimate the molecular weight of the dystrophin protein in the corrected clones. Expected molecular weight: 420 kDa for the reading-frame-corrected clone, 414 kDa for the exon-skipping clone, and 427 kDa for the exon 44 knockin clones and healthy control. An anti-dystrophin C terminus (amino acids 3661–3677) antibody was used to detect dystrophin protein, and an anti-α-SMA antibody was used as the sample loading control.
