Gong et al. report that more than one-third of human bromodomain (BRD)-containing proteins change localization in response to DNA damage. They identified ZMYND8 as a novel DNA damage response factor that recruits the nucleosome remodeling and histone deacetylation (NuRD) complex to damaged chromatin to repress transcription and promote repair by homologous recombination.
Keywords: DNA damage, chromatin, histone acetylation, bromodomain, ZMYND8
Abstract
How chromatin shapes pathways that promote genome–epigenome integrity in response to DNA damage is an issue of crucial importance. We report that human bromodomain (BRD)-containing proteins, the primary “readers” of acetylated chromatin, are vital for the DNA damage response (DDR). We discovered that more than one-third of all human BRD proteins change localization in response to DNA damage. We identified ZMYND8 (zinc finger and MYND [myeloid, Nervy, and DEAF-1] domain containing 8) as a novel DDR factor that recruits the nucleosome remodeling and histone deacetylation (NuRD) complex to damaged chromatin. Our data define a transcription-associated DDR pathway mediated by ZMYND8 and the NuRD complex that targets DNA damage, including when it occurs within transcriptionally active chromatin, to repress transcription and promote repair by homologous recombination. Thus, our data identify human BRD proteins as key chromatin modulators of the DDR and provide novel insights into how DNA damage within actively transcribed regions requires chromatin-binding proteins to orchestrate the appropriate response in concordance with the damage-associated chromatin context.
Endogenous and exogenous factors are constantly threatening genome integrity (Jackson and Bartek 2009). DNA double-strand breaks (DSBs) are particularly dangerous and can promote genome instability, a hallmark of cancer (Aguilera and Garcia-Muse 2013). Cells respond to these genetic insults by mounting the DNA damage response (DDR), which includes a network of proteins that function collectively to detect, signal, and repair the lesion (Jackson and Bartek 2009; Ciccia and Elledge 2010; Aguilera and Garcia-Muse 2013). Many DDR activities take place within chromatin, the protein–DNA complex that organizes the eukaryotic nuclear genome and regulates both epigenome and genome functions (Miller and Jackson 2012; Aguilera and Garcia-Muse 2013). Upon DNA damage, the DDR must coordinate responses to repair damage regardless of the genome or epigenome context. For example, the repair of DSBs by the two main pathways—homologous recombination (HR) and nonhomologous end-joining (NHEJ)—do not act stochastically across the genome (Chapman et al. 2012). Indeed, DSBs within active genes are preferentially repaired by HR (Aymard et al. 2014). Genes near DSBs are transcriptionally silenced by the DDR kinases ATM (Shanbhag et al. 2010) and DNA-PK (Pankotai et al. 2012). Thus, chromatin and DDR factors must cooperate to mount the requisite response for repairing DSBs within any chromatin setting, which can be highly variable throughout the genome. Understanding chromatin-based mechanisms that promote the DDR remains a critical question, as genome and epigenome alterations can severely compromise normal cell homeostasis.
Histone post-translational modifications (PTMs) are integral chromatin components of DNA damage signaling and repair (Miller and Jackson 2012). For example, γH2AX (i.e., phosphorylated H2AX on Ser139), is induced at DSBs and recruits the DDR protein MDC1 to damaged chromatin. This event promotes accumulation of many DDR factors at DSBs that orchestrate several DDR functions (Polo and Jackson 2011; Chen et al. 2013). Transcription-associated H3K36 methylation promotes DNA repair (Aymard et al. 2014; Carvalho et al. 2014; Jha and Strahl 2014; Pai et al. 2014; Pfister et al. 2014). PTMs can act together, as both methylated H4K20 and H2A ubiquitination are bound by the DDR factor 53BP1 at damaged chromatin (Fradet-Turcotte et al. 2013).
Acetylated histones are implicated in the DDR, as several histone acetyltransferases (HATs) and histone deacetylases (HDACs) are recruited to DNA damage and promote DNA damage signaling and repair (Gong and Miller 2013). Histone acetylation can alter DNA and histone binding, which is exemplified by H4K16 acetylation, which interferes with histone interactions to promote chromatin relaxation (Shogren-Knaak et al. 2006). TIP60 promotes HR by acetylating H4K16, which interferes with 53BP1 chromatin binding, which reduces NHEJ, a pathway antagonized by HDAC1 and HDAC2 (Miller et al. 2010; Tang et al. 2013). PTMs are recognized by “reader” domains that bind specific histone marks (Musselman et al. 2012). The primary reader domain for acetyl-lysines is the bromodomain (BRD). There are 42 human BRD-containing proteins (Filippakopoulos et al. 2012). Generally classified as transcriptional regulators, several BRD proteins (e.g., p300, TRIM28, BAZ1B, and BRD4) are involved in the DDR (Barnett and Krebs 2011; Iyengar and Farnham 2011; Ogiwara et al. 2011; Floyd et al. 2013). BRD proteins are potential candidates for reading DDR-specific acetylation changes occurring at DNA damage sites. This notion, along with the therapeutic success of inhibitors targeting the BRD of the BET family (e.g., BRD4) (Dawson et al. 2012; Filippakopoulos and Knapp 2014) and the development of DDR-targeting anti-cancer therapies (Helleday et al. 2008), warrants a detailed analysis of BRD proteins in the DDR.
As DDR factors often change localization upon DNA damage, we surveyed the mobilization of BRD proteins in response to DNA damage by laser microirradiation. We identified 14 BRD proteins that respond to DNA damage, including 12 that are recruited to DNA damage. We identify and provide mechanistic insights into the function of a novel DDR factor, ZMYND8 (zinc finger and MYND [myeloid, Nervy, and DEAF-1] domain containing 8). Our data revealed that ZMYND8 binds acetylated damaged chromatin, including actively transcribed regions, as a means of recruiting the nucleosome remodeling and histone deacetylation (NuRD) chromatin remodeling complex for transcriptional repression and DSB repair by HR. Collectively, these findings provide a comprehensive survey of BRD proteins in the DDR and demonstrate the functional importance of this protein family in reading chromatin signals to mediate chromatin-based responses to DNA damage in human cells.
Results
Dynamics of BRD proteins upon DNA damage
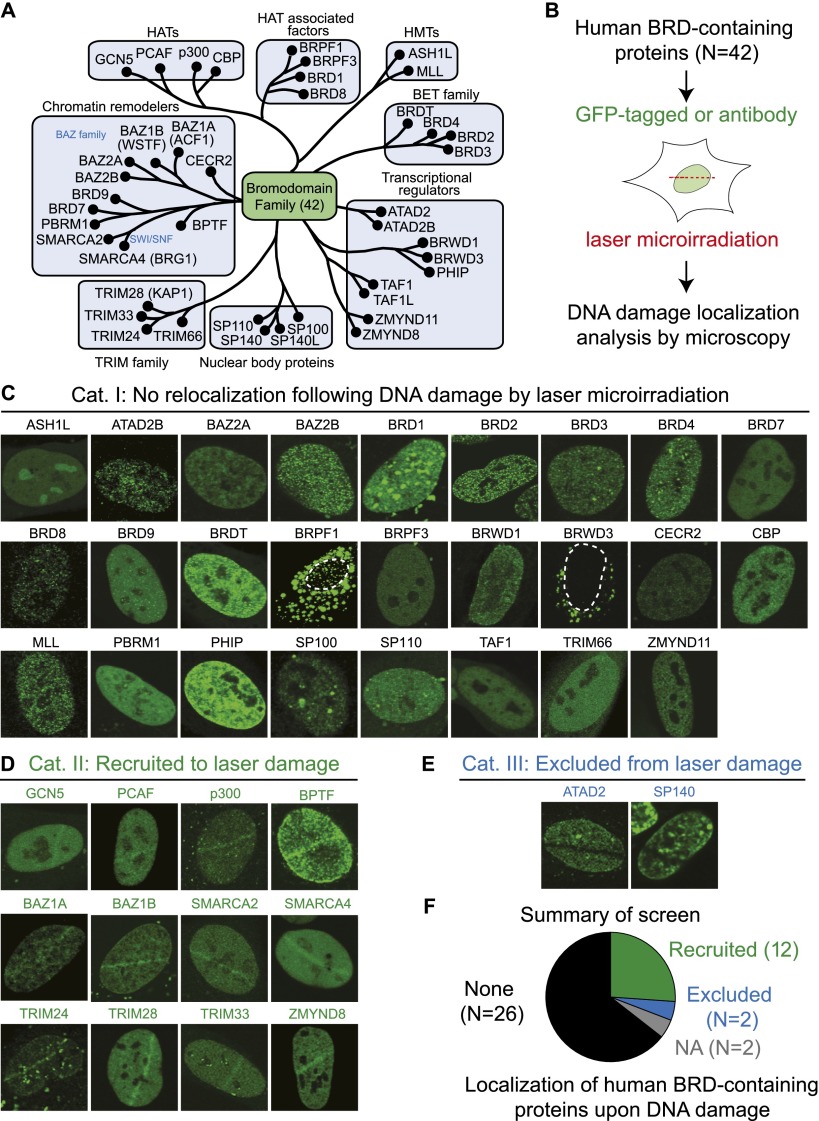
Human BRD proteins are involved in several nuclear functions, including transcription (Fig. 1A). DDR factors often exhibit dynamic mobilization following DNA damage. Therefore, we used laser microirradiation to study BRD protein localization in response to DNA damage. Laser microirradiation creates localized tracks of DNA damage in cells where protein dynamics can be monitored in live or fixed cells by fluorescence microscopy (Fig. 1B). We cloned 32 full-length GFP-tagged BRD proteins and acquired eight specific antibodies against other BRD proteins when cloning was unsuccessful. These reagents covered nearly all human BRD proteins, allowing us to analyze the response of this family to DNA damage (Fig. 1B).
Figure 1.
Comprehensive screening of human BRD protein relocalization following DNA damage. (A) Human BRD protein family, organized by known functions and the classifications from Filippakopoulos et al. (2012). (B) DNA damage relocalization screen for BRD proteins. (C–E) Screening results were sorted into three categories. (C) Category I: no relocalization following DNA damage. (D) Category II: recruited to DNA damage. (E) Category III: excluded from DNA damage. (F) Summary of screen results. (NA) Not analyzed.
Our screen identified three categories of responses to DNA damage (categories I–III). Category I contained 26 BRD proteins that did not respond to DNA damage (Fig. 1C). This analysis provided information about the normal subcellular localization of these proteins and acted as a negative control. Indeed, 25 out of 26 BRD proteins in category I were nuclear, and, although several exhibited focal nuclear accumulations, these were unresponsive to laser damage (e.g., BRD1, BRD3, BRPF1, and SP100) (Fig. 1C). We conclude from our results that most BRD proteins do not relocalize upon DNA damage.
Category II contained 12 BRD proteins that were recruited to DNA damage (Fig. 1D). These results confirmed studies identifying DNA damage recruitment of GCN5 (Guo et al. 2011), p300 (Ogiwara et al. 2011), BAZ1A (Lan et al. 2010), SMARCA2 (Ogiwara et al. 2011), SMARCA4 (Park et al. 2006), TRIM28 (Ziv et al. 2006), and TRIM33 (Kulkarni et al. 2013). Validation of these studies corroborated our method for identifying DNA damage recruitment of BRD proteins. However, there were some BRD proteins, including CBP and PBRM1, that are recruited to DNA damage but were not identified in our screen (Ogiwara et al. 2011; Kakarougkas et al. 2014). We also note that we identified SMARCA4 in our screen but not BRD7 or PBRM1, two proteins that are also members of complexes that contain SMARCA4 (Wilson and Roberts 2011). There are several explanations for these discrepancies, including the use of different experimental conditions and/or reagents. Many chromatin proteins reside in multiple complexes, which can complicate the conclusions gained from these results. Regardless, we identified five new BRD proteins (PCAF, BPTF, BAZ1B, TRIM24, and ZMYND8) that accrued at damage sites (Fig. 1D).
While DDR factors are often recruited to DNA damage, some are disassembled from damage, including the RNA processing factor THRAP3 (Beli et al. 2012). Interestingly, we identified ATAD2 and SP140 as BRD proteins excluded from laser damage (Fig. 1E). In summary, we found 14 DNA damage-responsive BRD proteins, including 12 that were recruited to DNA damage, and two that were excluded (Fig. 1F). Taken together, our screen revealed a large collection of BRD proteins that responded to DNA damage, suggesting an important involvement of this protein family in the DDR.
ZMYND8 DNA damage recruitment is BRD-dependent
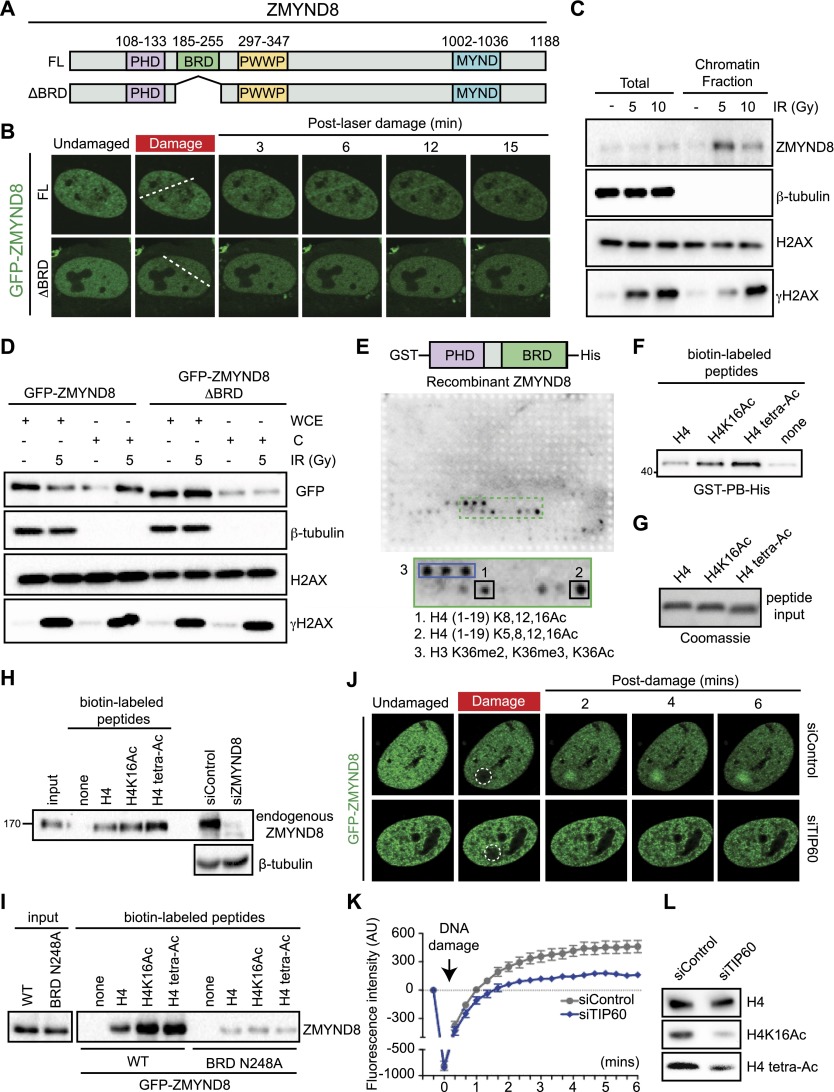
Although several BRD proteins accumulate at damage, the requirement of the BRD in this response is poorly understood. We sought to identify proteins that required the BRD for recruitment to damaged chromatin. After testing several candidates, we found that ZMYND8 required the BRD for damage recruitment (Fig. 2A,B; Supplemental Fig. S1A,B). Little is known about the function of ZMYND8, but it has been linked to transcription regulation (Zeng et al. 2010; Malovannaya et al. 2011). ZMYND8 contains a plant homeodomain (PHD), a BRD, a Pro–Trp–Trp–Pro (PWWP) chromatin-binding domain, and a protein–protein interaction MYND domain (Fig. 2A). Ectopically expressed GFP-ZMYND8 rapidly accumulated at laser damage, which was dependent on the BRD (Fig. 2B; Supplemental Fig. S1C). These results were corroborated with another method, as endogenous or GFP-tagged ZMYND8 increased chromatin association following DSB induction by ionizing radiation (IR) (Fig. 2C,D). Consistent with live-cell imaging, deletion of the ZMYND8 BRD decreased its chromatin association following IR (Fig. 2D). Thus, these results put forward ZMYND8 as a new DDR factor reliant on its BRD for binding damaged chromatin.
Figure 2.
TIP60-mediated H4 acetylation recruits ZMYND8 to damaged chromatin through BRD recognition. (A) ZMYND8 domain organization, including the PHD, BRD, PWWP domain, and MYND domain. (B) Laser damage recruitment of GFP-tagged full-length (FL) but not BRD-deleted (ΔBRD) ZMYND8. The dotted line indicates the laser path. (C) Endogenous ZMYND8 accumulates on chromatin following DNA damage by IR. Cells treated and analyzed as in D with the indicated antibodies. (D) Chromatin association of GFP-ZMYND8 following IR is BRD-dependent. Whole-cell extract (WCE) and chromatin (C) fractions obtained from untreated or IR-treated cells and analyzed by Western blotting. (E) Recombinant ZMYND8 PHD–BRD (GST-PB-His) binds H4Ac. Binding assays with the histone peptide arrays were performed as described in the Materials and Methods. Highly bound peptides are indicated. (Blue box) Modified H3K36 peptides. (F) Validation of the H4Ac interaction from E by peptide pull-down assay. Pull-down of recombinant ZMYND8 by the indicated peptides. (G) Coomassie staining of the loading control for H4 peptides. (H) Endogenous ZMYND8 from HeLa nuclear extracts binds H4Ac peptides. Extracts from siRNA-treated cells serves as control for ZMYND8 antibody specificity. β-Tubulin acted as a loading control. (I) GFP-ZMYND8 H4Ac binding requires the BRD. Experiments performed as in H using HEK293T cell extracts. A single N248A mutation within a conserved BRD acetyl-lysine-binding site reduces H4Ac interactions. (J) TIP60 depletion impairs ZMYND8 damage associations. U2OS cells stably expressing GFP-ZMYND8 and treated with siControl or siTIP60 were damaged within the dotted circles and imaged by live-cell microscopy. (K) Quantification of J. The difference in average fluorescence intensity of GFP-ZMYND8 in damaged versus undamaged regions is plotted at each time point. Error bars indicate SEM; n > 10. (L) TIP60 depletion reduces H4Ac. Extracts from siControl and siTIP60 cells were analyzed by Western blotting with the indicated antibodies. (H4 tetra-Ac) Acetylation of H4 at K5, K8, K12, and K16.
The ZMYND8 BRD binds TIP60-mediated acetylated H4 (H4Ac) for DNA damage recruitment
Since ZMYND8 required the BRD for damage association, we set out to identify the molecular determinants of ZMYND8 chromatin binding. Tandem PHD and BRD configurations are observed in chromatin proteins and can contribute to cooperative recognition of histone PTMs (Tsai et al. 2010; Ruthenburg et al. 2011). Using modified histone peptide arrays, we analyzed recombinant ZMYND8 PHD–BRD binding to histone modifications (Fig. 2E). This analysis identified ZMYND8 PHD–BRD-binding H4Ac. Pull-down assays with unmodified or H4Ac peptides confirmed these interactions and revealed a binding preference for H4Ac (Fig. 2F). We obtained identical results for full-length endogenous and GFP-tagged ZMYND8 using peptide pull-down assays with HeLa nuclear extracts and cells expressing GFP-ZMYND8, respectively (Fig. 2G–I). The observed interaction between ZMYND8 and H4Ac prompted us to ask whether this interaction was BRD-dependent. BRDs contain several key conserved residues that mediate acetyl-lysine binding (Filippakopoulos et al. 2012). Mutation of one of these residues in the BRD of ZMYND8, N248A, reduced binding to H4Ac peptides (Fig. 2I). Several BRD proteins also contain a PHD that is organized similarly to ZMYND8 (Fig. 2A; Filippakopoulos et al. 2012). However, the presence of a PHD–BRD structure does not predict the involvement of a protein in the DDR or a binding preference for H4Ac. For example, TRIM28 and BAZ1B both contain PHD–BRDs that are dispensable for DNA damage recruitment (Supplemental Fig. 1A,B). ZMYND11, a paralog of ZMYND8 that regulates transcription and impacts tumor suppression, also contains a PHD–BRD (Wen et al. 2014). Our analysis did not detect its recruitment to DNA damage or binding to H4Ac (Fig. 1C; Supplemental Fig. S2A). Taken together, we conclude that ZMYND8 binds H4Ac via a BRD-dependent interaction.
Acetylation of H4, including H4K16Ac, functions within the DDR (Gong and Miller 2013). The HAT TIP60 acetylates all N-terminal H4 lysine residues and plays diverse roles in both transcriptional regulation and the DDR (Ikura et al. 2000). Since TIP60 and ZMYND8 are both damage-recruited and TIP60 acetylates H4, a potential binding substrate for ZMYND8, we examined the role of TIP60 in ZMYND8 mobilization at damage sites by live-cell imaging. The rapid recruitment of ZMYND8 at damage sites was significantly reduced in TIP60-depleted cells (Fig. 2J, quantified in K; siRNA knockdown efficiency in Supplemental Fig. S2B), as were H4K16Ac and H4 tetra-Ac (acetylation of H4 at K5, K8, K12, and K16.) levels (Fig. 2L; Supplemental Fig. S2C). As a control, we depleted the HAT MOF, which targets histone H4 and is involved in the DDR (Sharma et al. 2010; Gong and Miller 2013). Depletion of MOF, unlike TIP60, did not reduce ZMYND8 recruitment to DNA damage (Supplemental Fig. S2D–G). Thus, ZMYND8 association with damaged chromatin specifically requires TIP60. The binding of ZMYND8 to H4Ac is consistent with this mark mediating the damage association of ZMYND8 with chromatin, although additional TIP60 targets could contribute to these effects.
ZMYND8 interacts with chromatin and chromatin-modifying complexes
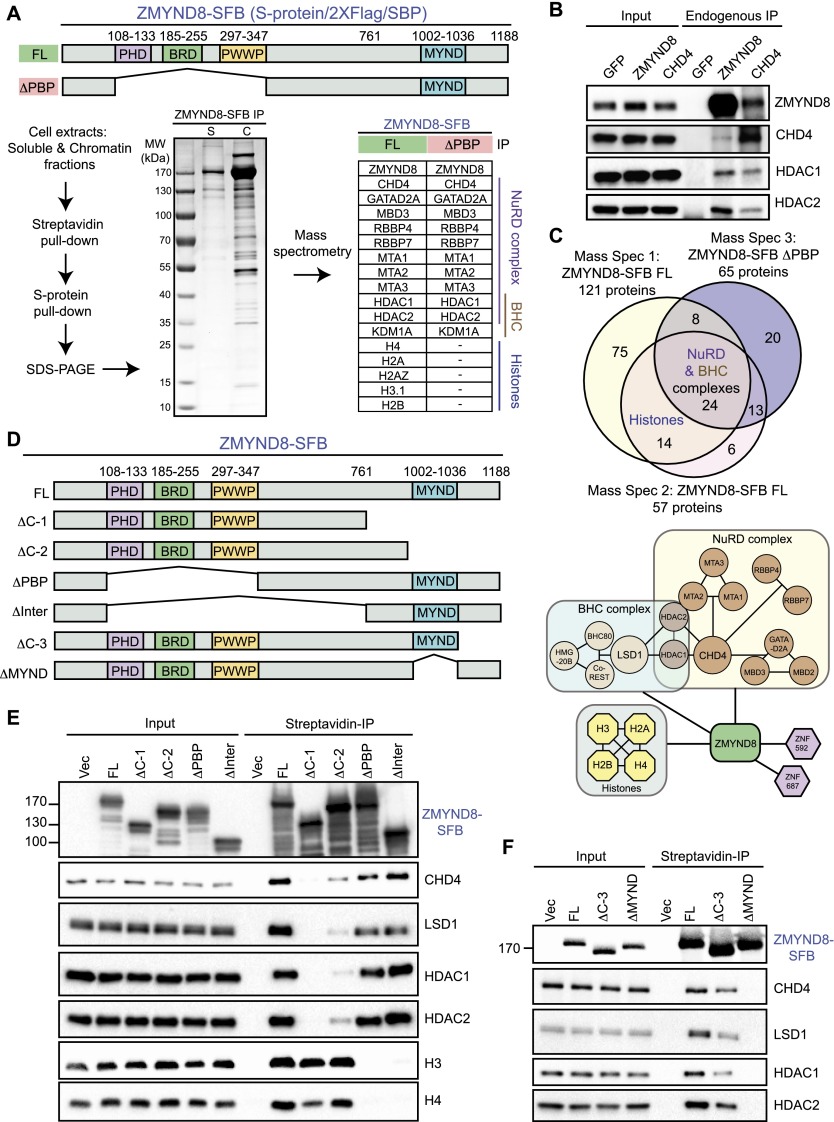
Proteomic studies have reported interactions between ZMYND8 and transcriptional regulators, including the estrogen receptor, but the functions of these interactions are unclear (Malovannaya et al. 2011; Eberl et al. 2013). To extend these analyses, we identified ZMYND8 interactors by mass spectrometry (MS) (Fig. 3A). Several independent experiments revealed an overlapping set of 38 ZMYND8-interacting proteins (Fig. 3C; Supplemental Table S1). These included members of the NuRD chromatin remodeling complex and the BRAF–HDAC (BHC) complex (Fig. 3A), confirming previous large-scale proteomic screens (Malovannaya et al. 2011; Eberl et al. 2013). We validated the NuRD interaction with reciprocal coimmunoprecipitation (co-IP) analysis of endogenous ZMYND8 and CHD4, a core catalytic component of NuRD (Fig. 3B). We also performed MS analysis on purified ZMYND8 lacking its chromatin interaction domains. In these purified complexes, histone, but not NuRD and BHC, interactions were lost (Fig. 3A,C). These findings demonstrate that while the PHD–BRD–PWWP chromatin-binding modules are responsible for ZMYND8 chromatin association, these domains are dispensable for interactions with the NuRD and BHC chromatin-modifying complexes. These results suggested that ZMYND8 could link chromatin-modifying complexes (e.g., NuRD and BHC) to chromatin, including in response to DNA damage.
Figure 3.
ZMYND8 interacts with chromatin and chromatin-modifying complexes NuRD and BHC. (A) Identification of ZMYND8-interacting factors by MS. Purification scheme of full-length (FL) and PHD–BRD–PWWP-deleted (ΔPBP) SFB-tagged ZMYND8. (SBP) Streptavidin-binding peptide. The table represents ZMYND8 interactors identified in mass spectrum from the indicated experiments. (B) Endogenous ZMYND8 interacts with CHD4, a core component of the NuRD complex. Western blotting analysis of reciprocal co-IPs with the indicated antibodies from HEK293T cells. (C) ZMYND8 MS data. Overlap of full-length (n = 2) and ΔPBP ZMYND8 interactors. (Bottom) Individual interactions based on literature and our MS results. (D–F) The BRD of ZMYND8 interacts with chromatin, and the MYND domain interacts with the NuRD and BHC complexes. (D) Full-length and mutant SFB-ZMYND8 constructs. (E,F) Mapping ZMYND8 interaction domains with interactors. ZMYND8 constructs were transfected into HEK293T and analyzed by co-IP. Full-length and mutant SFB-ZMYND8 were purified using streptavidin beads. (input) Whole-cell extracts. Purified complexes and input were analyzed by Western blotting.
To address this question, we mapped ZMYND8 regions that interacted with NuRD and BHC complexes. We created and expressed a series of ZMYND8 deletion mutants for immunoprecipitation interaction studies in cells (Fig. 3D). The NuRD and BHC complex members, including CHD4 and LSD1, respectively, coimmunoprecipitated with ZMYND8 (Fig. 3E). Consistent with MS analysis, deletions of PHD–BRD–PWWP abolished interactions with chromatin (i.e., H3 and H4) while having little effect on NuRD and BHC associations (Fig. 3E). Conversely, deletions within the C terminus that included the MYND domain reduced ZMYND8 interactions with chromatin-modifying complexes but not chromatin (Fig. 3E). Deletion of the MYND domain of ZMYND8 abolished interactions with the NuRD and BHC complexes (Fig. 3F). Thus, these findings identify the MYND domain as the region of ZMYND8 that interacts with the NuRD and BHC complexes, while ZMYND8 PHD–BRD–PWWP domains are responsible for chromatin binding.
ZMYND8 recruits CHD4 to damaged chromatin
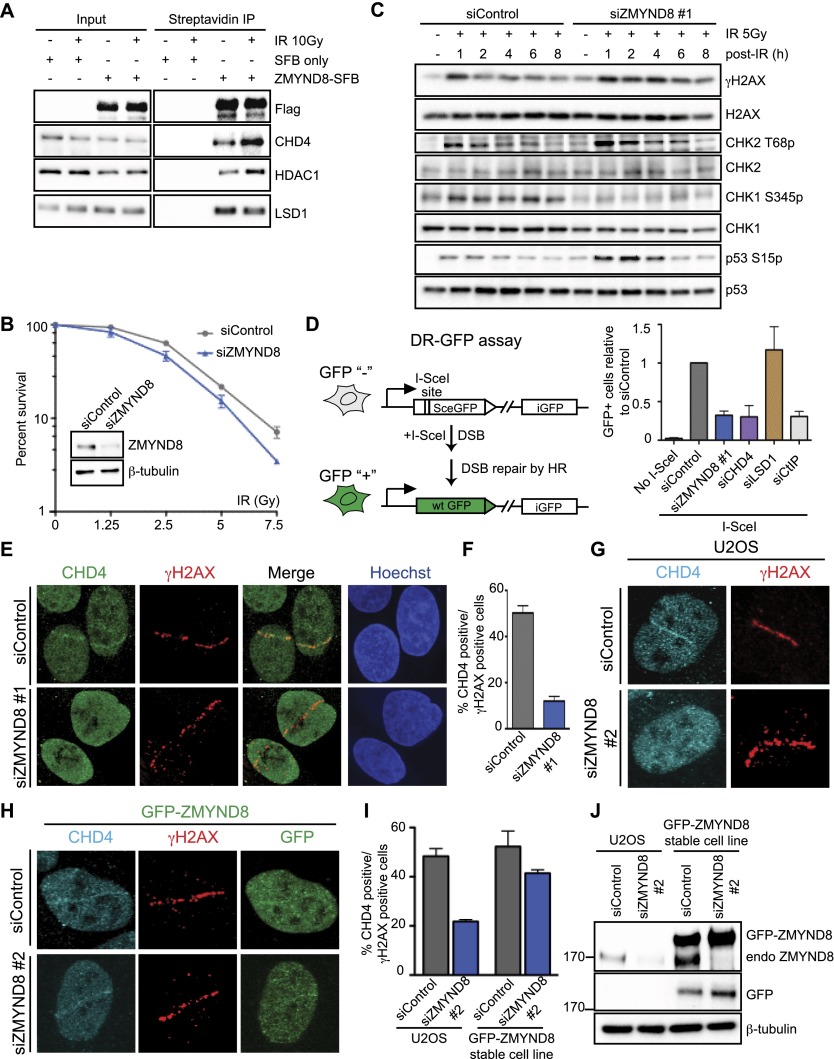
To further explore the functional relationship between ZMYND8, NuRD, and BHC complexes in the DDR, we analyzed these interactions after DNA damage. We identified these complexes in ZMYND8 immunoprecipitations following DNA damage by MS, along with several other DNA damage-specific interactors (Supplemental Fig. S3). Additional co-IP analysis revealed that CHD4, but not LSD1, increased their association with ZMYND8 following DNA damage (Fig. 4A). Given our validated ZMYND8 interactions with the NuRD component CHD4 and the BHC complex member LSD1, we focused on exploring these interactions in the DDR.
Figure 4.
ZMYND8 participates in the DDR and recruits CHD4 to damaged chromatin. (A) ZMYND8 interacts with CHD4 upon IR treatment. Experiments were performed as in Figure 3E with IR. (B) Clonogenic assays reveal hypersensitivity of ZMYND8-depleted cells to IR. U2OS cells were treated with control or ZMYND8 siRNAs and damaged with various doses of IR. Graphs are mean ± SEM; n = 2. (C) ZMYND8-depleted cells are defective in DNA damage signaling. siControl and siZMYND8 U2OS cells were IR-treated and analyzed at the indicated time points by Western blotting. Several phosphorylated DNA damage markers were analyzed with unmodified antibodies acting as loading controls. (D) Cells depleted of ZMYND8 and CHD4, but not LSD1, are defective in HR. DR-GFP reporter assays were performed after depletion of the indicated proteins by siRNAs. Depletion of CtIP was used as a positive control. Error bars indicate SEM; n = 3. (E–G) Recruitment of CHD4 to laser damage requires ZMYND8. siControl and siZMYND8 U2OS cells were laser-damaged, and endogenous CHD4 accumulation was analyzed by immunofluorescence. (F) Quantification of E. Data were obtained from >50 cells from three independent experiments. Error bars indicate mean ± SEM. (G) The same results obtained as in E using an independent siRNA targeting the 3′ untranslated region (UTR) of ZMYND8. (H–J) Ectopically expressed GFP-ZMYND8 rescues defective CHD4 damage accrual in ZMYND8-depleted cells. U2OS cells stably expressing GFP-ZMYND8 were treated with si3′ UTR targeting endogenous but not GFP-tagged ZMYND8, which lacks the 3′ UTR. (I) Quantification of G and H performed as in F. n = 2. (J) Western blot analysis of samples from G and H with the indicated antibodies. Note: 3′ UTR siRNA targeting ZMYND8 depletes endogenous but not GFP-tagged ZMYND8.
Our identification of ZMYND8 as a factor that accumulated on damaged chromatin suggested an involvement in the DDR. Consistent with this idea, depletion of ZMYND8 caused hypersensitivity to the DSB-inducing agent IR (Fig. 4B; Supplemental Fig. S4A). Furthermore, while IR induced the phosphorylation of several DSB markers—including γH2AX, p53-S15, and CHK2-T68 (Fig. 4C)—these were sustained in ZMYND8-depleted cells, suggesting DSB repair deficiencies in these cells. Interestingly, these are similar to defects observed in CHD4-deficient cells (Larsen et al. 2010; Smeenk et al. 2010). Conversely, CHK1 phosphorylation, a modification linked with DNA end resection and HR, was reduced in ZMYND8-deficient cells (Fig. 4C). These results were not merely due to lower CHK1 transcription or CHK1 protein levels, which were not significantly different between siControl and siZMYND8 cells (Supplemental Fig. 4B,C). Collectively, these data pointed toward a function for ZMYND8 in the DDR and furthermore suggested that these effects could be mediated through CHD4.
As CHD4 is involved in DSB repair by HR (Pan et al. 2012), we used a HR reporter assay to test the role of ZMYND8 in HR (Pierce et al. 1999). ZMYND8 depletion reduced HR using this assay (Fig. 4D; Supplemental Fig. S4D). We also confirmed results from previous studies, as depletion of CHD4 but not LSD1 reduced HR (Fig. 4D; Pan et al. 2012; Mosammaparast et al. 2013). Flow cytometry, EdU incorporation, and proliferation analyses revealed little change in cell cycle distribution or growth rates in siZMYND8 cells compared with siControl cells, thus ruling out any potential impact of cell cycle changes on these results (Supplemental Fig. S5A–E). These findings strongly indicated ZMYND8 in DSB repair by HR. We considered that ZYMDN8 and CHD4 could act in the same pathway for HR, as we observed increased interactions between these factors after DNA damage, and they exhibited similar DNA damage signaling and repair phenotypes upon their depletion. To explore this possibility, ZMYND8 recruitment to laser damage was studied in CHD4- and LSD1-depleted cells. Depletion of either CHD4 or LSD1 did not affect ZMYND8 accumulation at damage (Supplemental Fig. S6A–C). CHD4 and LSD1 are recruited to DNA damage (Chou et al. 2010; Larsen et al. 2010; Polo et al. 2010; Smeenk et al. 2010; Mosammaparast et al. 2013). To address whether ZMYND8 could act upstream or independently of these factors, CHD4 and LSD1 recruitment to damage sites was analyzed in ZMYND8-depleted cells. Strikingly, CHD4 accumulation at damage sites was reduced in ZMYND8 knockdown cells, while LSD1 was unaffected (Fig. 4E,F; Supplemental Fig. 6C,D). We confirmed ZMYND8-dependent CHD4 DNA damage recruitment using an independent siRNA targeting the 3′ untranslated region (UTR) of ZMYND8 (Fig. 4G). Furthermore, CHD4 recruitment defects in ZMYND8-depleted cells were rescued in cells stably expressing a siRNA-resistant GFP-tagged ZMYND8 (Fig. 4H, quantified in I, note that ZMYND8 but not GFP-ZMYND8 protein levels are reduced by ZMYND8 3′ UTR siRNA, as shown in J). Collectively, these results rule out that our observations were due to siRNA off-target effects. Thus, these findings demonstrate a role for ZMYND8 in DNA damage signaling and repair by recruiting the NuRD complex to damaged chromatin.
ZMYND8 requires active transcription for damage recruitment to promote HR
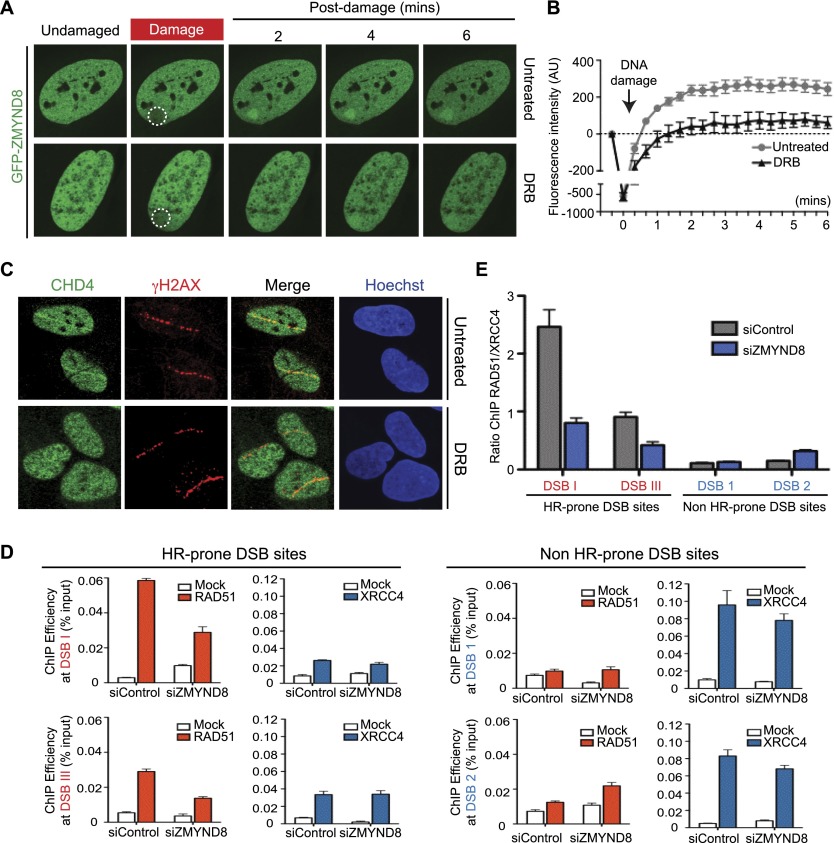
We next addressed how ZMYND8 accrued on damaged chromatin. We speculated that active transcription could control this event, as ZMYND8 bound H4Ac, a mark associated with active transcription and DSBs (Price and D’Andrea 2013). Strikingly, transcription inhibition by the RNA polymerase II (RNAP II) inhibitor DRB (5,6-dichlorobenzimidazole riboside) abolished the accumulation of ZMYND8 at DNA damage sites (Fig. 5A, quantified in B). CHD4 recruitment to damage was also reduced by transcriptional inhibition (Fig. 5C; quantified in Supplemental Fig. S7G). This effect appeared relatively specific for ZMYND8 and CHD4, as recruitment of other BRD proteins were unaffected by DRB treatment (Supplemental Fig. S7A–F). Inhibition of nascent transcription was confirmed, as 5-ethynyl uridine (EU) labeling was impeded by DRB treatment (Supplemental Fig. S7H; Beli et al. 2012; Adam et al. 2013). We conclude from these data that ZMYND8 and NuRD require transcriptionally active chromatin to mediate their recruitment to DNA damage.
Figure 5.
ZMYND8 identifies damage within transcriptionally active chromatin to promote HR. (A) Recruitment of ZMYND8 to damaged chromatin requires active transcription. Cells were analyzed as in Figure 2J with or without treatment with the transcriptional inhibitor DRB. (B) Quantification of A. Error bars indicate SEM; n > 10. (C) Recruitment of CHD4 to damaged chromatin requires active transcription. Cells were analyzed as in Figure 4E with or without DRB treatment. (D,E) ZMYND8 promotes RAD51 loading at HR-prone DSB sites within active chromatin. Samples from DlvA cells containing site-specific AsiSI-induced DSBs were analyzed by ChIP analysis for the HR factor RAD51 or NHEJ factor XRCC4 as described previously (Aymard et al. 2014). DSB I and DSB III represent HR-prone DSB sites, while DSB 1 and DSB 2 represent Non-HR-prone DSB sites. ChIP efficiency was measured by percentage of input, and mock versus antibody-containing data are graphed for each DSB from siControl and siZMYND8 #1 samples. (E) RAD51/XRCC4 ratios of data obtained in D.
DSBs within transcriptionally active chromatin are preferentially repaired by HR, and transcription inhibition reduces HR factor loading at such sites (Aymard et al. 2014). ZMYND8 was a good candidate for promoting HR within transcriptionally active loci, as ZMYND8 depletion reduced HR (Fig. 4D), and its damage recruitment required active transcription (Fig. 5A). To explore this possibility, we used a system developed by the Legube laboratory (Aymard et al. 2014) in which sequence-specific DSBs are induced at transcriptionally active and inactive loci by the AsiSI restriction enzyme. At those breaks, repair factor loading of RAD51 or XRCC4 strongly correlates with repair by HR or NHEJ, respectively (Aymard et al. 2014). As expected, chromatin immunoprecipitation (ChIP) analysis revealed an increased recruitment of RAD51 at HR-prone DSBs within transcriptionally active loci compared with non-HR-prone DSBs induced in transcriptionally inactive regions (Fig. 5D). Remarkably, ZMYND8 depletion impaired RAD51 recruitment at HR-prone DSBs but not at non-HR-prone DSBs, while reduced ZMYND8 levels had no effect on XRCC4 (i.e., NHEJ) loading at either DSB site category (Fig. 5D). Accordingly, ZMYND8 depletion led to a strong decrease of the RAD51/XRCC4 enrichment ratio at HR-prone DSBs, indicative of an impaired use of HR at DSBs induced in transcriptionally active regions (Fig. 5E). These results are consistent with the depletion of ZMYND8 reducing HR repair (Fig. 4D) but not affecting NHEJ (Supplemental Fig. S8A,B). Taken together, we conclude that ZMYND8 promotes HR, including within transcriptionally active chromatin.
ZMYND8 mediates transcriptional silencing within damaged chromatin
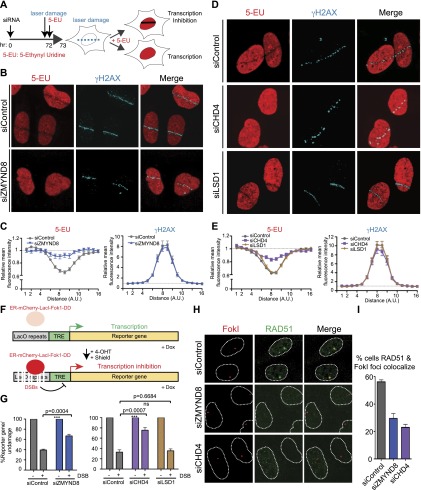
DNA damage inhibits transcription locally (Chou et al. 2010; Iacovoni et al. 2010; Shanbhag et al. 2010; Pankotai et al. 2012; Soria et al. 2012; Adam et al. 2013), and the DDR regulates this event to avoid conflicts between transcription and repair activities, which can threaten genome–epigenome integrity (Svejstrup 2010; Adam and Polo 2014). Since ZMYND8 was damage-recruited and promoted HR at transcriptionally active damaged sites, we tested whether ZMYND8 repressed transcription after damage. Using EU labeling to monitor nascent transcription (Fig. 6A), we found that ZMYND8-depleted cells exhibited reduced transcriptional silencing at damaged sites (Fig. 6B, quantified in C). These results were confirmed using an independent ZMYND8 siRNA and by rescuing this defect in cells expressing a siRNA-resistant ZMYND8, which ruled out any siRNA off-target effects (Supplemental Fig. S9A–D). We also observed that TIP60 was required for transcriptional repression following DNA damage, consistent with our observation that ZMYND8 recruitment to damage sites is reliant on TIP60 (Supplemental Fig. S10A,B). Consistent with ZMYND8 damage recruitment of CHD4, depletion of CHD4 but not LSD1 resulted in defective transcriptional repression after DNA damage (Fig. 6D, quantified in E). Thus, ZMYND8 and the NuRD complex promote transcriptional repression after DNA damage, extending previous results implicating NuRD in this process (Chou et al. 2010). Consistent with transcriptional repression inhibiting ZMYND8 recruitment, we observed transient ZMYND8 association with DNA damage. Indeed, ZMYND8 accumulation at laser damage was observed at 15 min but was nearly undetectable 25 min after DNA damage (Supplemental Fig. S10E). The DNA damage recruitment dynamics of ZMYND8 are similar to those reported for the NuRD complex (i.e., CHD4) (Polo et al. 2010). Using a system developed by the Greenberg laboratory (Tang et al. 2013) to monitor transcriptional repression specifically at DSBs, we observed that the depletion of ZMYND8 or CHD4 resulted in defective transcriptional repression upon DSB induction (Fig. 6F,G). Consistent with our findings that ZMYND8 and CHD4 were required for optimal RAD51 loading and HR using several experimental systems, we observed similar results using the FokI-induced DSB system (Shanbhag et al. 2010; Tang et al. 2013). Indeed, depletion of either ZMYND8 or CHD4 reduced RAD51 loading at FokI-mediated DSBs (Fig. 6H, quantified in I). Taken together, these results demonstrate that ZMYND8 and the NuRD complex promote transcriptional silencing at DSBs as well as facilitate HR within these DNA damage sites.
Figure 6.
ZMYND8 and CHD4 mediate transcriptional repression upon DNA damage. (A) Scheme of nascent transcription analysis by 5-ethynyl uridine (5-EU) monitoring following laser damage. (B) ZMYND8 promotes transcriptional repression following laser damage. Cells treated with control or ZMYND8 siRNAs were subjected to the scheme shown in A and analyzed by immunofluorescence. γH2AX marks DNA damage. (C) Quantification of 5-EU and γH2AX fluorescence intensity from B. Measurements of fluorescent intensity along lines perpendicular to the laser damage, which contained both damaged and undamaged regions, were obtained. Values were normalized to undamaged regions. Error bars indicate SEM; n > 10. (D) CHD4 but not LSD1 is required for transcriptional repression following laser damage. Experiments performed as in B. (E) Quantification of D as in C. (F) Scheme of U2OS DSB reporter cells adapted by permission from Macmillan Publishers Ltd. from Tang et al. (2013), © 2013. Shield-1 and 4-OHT regulate the Fok1 nuclease, which induces DSBs upstream of reporter genes within LacO repeats. Doxycycline induces transcription of the reporter gene, allowing transcriptional repression upon DSB induction to be measured by quantitative PCR (qPCR). (G) ZMYND8 and CHD4, but not LSD1, regulate transcriptional repression at DSBs. The system from F was analyzed in cells treated with the indicated siRNAs. Error bars indicate SEM; n = 4. P-values were calculated using Student’s t-test. (H) ZMYND8 and CHD4 promote RAD51 loading at DSBs. RAD51 loading by immunofluorescence in siControl, siZMYND8, and siCHD4 cells was analyzed 3 h after DSB induction using a FokI-inducible DSB system (Tang et al. 2013). (I) Quantification of H. Error bars indicate SEM; n = 3. siRNA #1 was used for siZMYND8 treatments.
Discussion
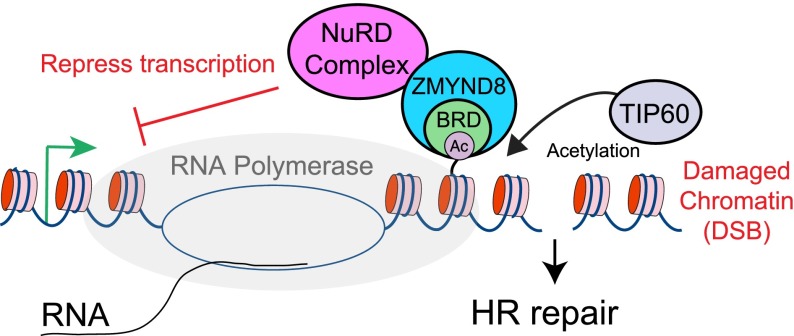
Our survey of BRD protein localization dynamics in response to DNA damage provides a cell-biological and broad examination of BRD chromatin reader proteins in the DDR (Fig. 1). Indeed, we identified 14 BRD proteins that assemble/disassemble at DNA damage sites. Among these DNA damage-recruited BRD proteins was ZMYND8, a new DDR factor that our mechanistic studies revealed is responsible for recruiting the NuRD chromatin remodeling complex to damaged chromatin to repress transcription and facilitate DNA repair by HR (Fig. 7).
Figure 7.
Model of DNA damage recognition pathway by ZMYND8. Upon DNA damage, ZMYND8 interacts with the NuRD complex. ZMYND8 recognizes TIP60-dependent acetylations (e.g., H4Ac) with its BRD, including within actively transcribing damaged chromatin, resulting in recruitment of the NuRD complex to these damage sites. The TIP60-dependent recruitment of the NuRD complex promotes transcriptional silencing that facilitates repair by HR, including within transcriptionally active damaged chromatin.
BRD proteins as mediators of the DDR
Chromatin greatly impacts the DDR, since the detection, signaling, and repair of damaged DNA occur within the context of chromatin. Histone PTMs regulate chromatin structure and function in response to DNA damage. Indeed, several HAT and HDAC enzymes that regulate histone acetylation are recruited to DNA damage to modify chromatin to facilitate DDR activities (Gong and Miller 2013). Despite extensive studies of acetylation signaling in the DDR, a comprehensive view of how acetylated chromatin impacts the association of proteins within damaged chromatin to promote the DDR has remained incomplete.
BRD proteins are the primary readers of acetylated chromatin, and several studies as well as our data reported here suggest the importance of this protein family in the DDR (Park et al. 2006; Lan et al. 2010; Ogiwara et al. 2011; Lee et al. 2012; Floyd et al. 2013; Kulkarni et al. 2013). Our screen identified damage-associated BRD proteins representing four general classes of proteins: (1) HATs, (2) chromatin remodelers, (3) TRIM proteins, and (4) ZMYND8. In most cases, we identified multiple BRD proteins in each category. Different BRD proteins could interact with nonoverlapping DNA lesions across the genome or target different substrates at the same damage site. It will also be important to examine the nature of the damage recognition of these factors (i.e., BRD-dependent or BRD-independent, its acetylation targets, etc.) as well as DNA damage-induced interactions with other proteins. The identification of damage-associated BRD proteins provides a useful guide for delineating the role of BRD proteins in the DDR in human cells. Additional studies are needed to provide a clearer understanding of the specific contribution that the BRD provides for these proteins for their DDR functions. This remains an important but unanswered question that the results from our screen have begun to address.
We observed exclusion from DNA damage sites for the BRD proteins ATAD2 and SP140. Similar observations for the RNA processing factor THRAP3 upon DNA damage have been reported (Beli et al. 2012). HDACs are recruited to damage sites and are involved in the DDR (Miller et al. 2010; Gong and Miller 2013). Thus, histone deacetylation could signal the disassembly of BRD proteins by removing their requisite chromatin-binding site. For ATAD2 and SP140, exclusion from damage sites could promote transcriptional repression, since these proteins can activate transcription (Bloch et al. 1999; Ciro et al. 2009). The function of protein removal from damaged chromatin is poorly understood, but our results suggest that acetylation signaling could be involved.
Our study has broad implications for deciphering chromatin-based mechanisms that maintain genome integrity. Successful small molecule inhibitors against BRD4 in oncology have garnered immense enthusiasm for drug development targeting BRD proteins (Dawson et al. 2012; Filippakopoulos and Knapp 2014). DDR factors are also actively pursued as therapeutic targets (Helleday et al. 2008). Given our identification of many BRD proteins as damage-recruited factors, a better understanding of the relationship between BRD proteins and the DDR will assist in guiding these therapeutic strategies.
The role of ZMYND8 in the DDR
Our findings define a DDR pathway involving ZMYND8 that identifies damaged chromatin to promote transcriptional repression and repair by HR by recruiting the NuRD complex to DNA damage. In support of this idea, ZMYND8 and NuRD depletion result in similar defects in DNA damage signaling, transcriptional repression, and HR repair, further highlighting the functional relationship between ZMYND8 and the NuRD chromatin remodeling complex. We also demonstrate the requirement for the HAT TIP60 in promoting ZMYND8 recruitment to DNA damage. Furthermore, DNA damage-induced transcriptional repression required TIP60. Previous work demonstrated that the TIP60 HAT activity is stimulated by trimethylation of H3K9 by a complex containing the SUV39h1 methyltransferase along with KAP1 and HP1. (Sun et al. 2009; Ayrapetov et al. 2014). Although this histone mark is normally associated with heterochromatin, the finding that this mark is induced in damaged euchromatin explained how TIP60 could be activated in both euchromatin and heterochromatin. Thus, our data are consistent with a model in which the observed TIP60 damage-induced acetylation of chromatin, including H4K16 (Tang et al. 2013), promotes NuRD recruitment to DNA damage through acetylation recognition by ZMYND8 (Fig. 7). The activity of the NuRD complex would then repress transcription and promote HR. Although H3K9me3 is associated with transcriptionally inactive chromatin regions, its role in repressing transcription upon DNA damage has not been explored. It is interesting to note that the recruitment of SUV39H1 and the NuRD complex, including ZMYND8, is transient, and both of these complexes require PARP (Supplemental Fig. S10E; Polo et al. 2010; Ayrapetov et al. 2014). Further studies are needed to fully delineate the regulation and potential interactions between these chromatin-repressive complexes that function at DNA damage sites to regulate transcription and repair. Based on our data, we propose a model in which, upon DNA damage, TIP60 activation results in chromatin acetylation at DNA damage that is recognized by ZMYND8 to recruit the NuRD complex (Fig. 7). These complexes further modify chromatin and potentially other DDR and/or chromatin-associated factors to repress transcription and promote DNA repair by HR. This work highlights the vital role that chromatin plays in orchestrating the DDR within the context of chromatin.
DNA damage-induced transcriptional silencing, a critical process for maintaining genome–epigenome integrity, is a poorly understood response to DNA damage. Discovery of the chromatin factors mediating DSB repair choice and transcriptional silencing is paramount for understanding how these processes cooperate to promote the DDR within complex chromatin states throughout the genome. Our identification of TIP60-dependent ZMYND8 and NuRD complex functions within these processes extends our understanding of how HR repair is promoted at transcription-associated damage. Although multiple chromatin remodeling complexes are recruited to DNA damage sites (Fig. 1; Lans et al. 2012), the NuRD complex is the only chromatin remodeling complex that contains HDACs that can silence transcription (Lai and Wade 2011). The NuRD complex normally represses transcription through specific interactions with methylated DNA, transcription factors, or histones (Lai and Wade 2011). In the context of DNA damage, ZMYND8 provides a new DNA damage recruitment mechanism for the NuRD complex. This allows the NuRD complex to identify damaged acetylated chromatin to mediate its recruitment and subsequent transcriptional silencing. Interestingly, NuRD was shown to interact with the zinc finger protein ZNF827 to target this complex to telomeres in alternative lengthening of telomeres (ALT) cells to promote HR (Conomos et al. 2014). Taken together, chromatin-modifying enzymes can target damaged DNA within varied chromatin landscapes, a property essential for the DDR, as DSBs can occur anywhere within diverse genome and epigenome landscapes.
Our data are consistent with the need to have transcription-dependent DNA damage surveillance systems that identify damage within transcribing chromatin to repress active transcription and promote repair by HR, a pathway that we identified as being mediated by TIP60, ZMYND8, and the NuRD complex. However, transcriptional silencing by RNAP II inhibition alone is not sufficient to promote HR at actively transcribed damaged chromatin (Aymard et al. 2014). Conversely, we observed that defective DNA end resection, a process required for HR repair, did not affect DNA damage-induced transcriptional repression (Supplemental Fig. S10C,D). These findings suggest that transcriptional repression occurs independently of HR initiation. Therefore, the chromatin remodeling and HDAC activities of NuRD appear to be required for two mechanistically distinct responses at damaged chromatin, including silencing transcription and remodeling chromatin, both of which contribute to HR repair.
ZMYND8 has been implicated in cancer as a cutaneous T-cell lymphoma-associated antigen, as a fusion protein with the oncogene RELA in acute erythroid leukemia and with CEP250 in breast cancer (Eichmuller et al. 2001; Panagopoulos et al. 2013; Wada et al. 2014). Both of the ZMYND8 fusions reported remove the MYND domain, the NuRD complex interaction region that we identified in ZMYND8, suggesting that defective CHD4 chromatin recruitment could contribute to these pathologies. Additionally, high-frequency mutations in CHD4 have been identified in several cancers, including endometrial tumors in which 17% contained CHD4 mutations (Lai and Wade 2011; Le Gallo et al. 2012). In light of our work, it will be worth studying ZMYND8 and its interaction with NuRD in cancer. Our demonstration that ZMYND8 promotes HR and transcriptional silencing within actively transcribed chromatin suggests that defects in this pathway, including those in CHD4, could result in mutations or aberrant transcriptional responses within active genes. The success of targeting HR-defective tumors (for example, BRCA1 and BRCA2 mutations) with PARP inhibitors (Helleday et al. 2008) should be analyzed in NuRD-deficient settings, based on our findings that ZMYND8 and CHD4 promote HR.
Our study suggests a widespread involvement of BRD proteins in orchestrating chromatin-based mechanisms of the DDR to promote genome maintenance. Our findings also provide insights into therapeutic applications for this disease-relevant protein family. As the BRD represents the major reader domain for acetyl-lysine modifications, our characterization of the involvement of the BRD proteins in associating with damaged chromatin will aid in elucidating the function of acetylation-dependent signaling in the DDR and in human diseases, including cancer.
Materials and methods
Cell culture conditions and treatments
Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells stably expressing GFP-tagged BRD proteins were established and maintained in medium with 10 μg/mL blasticidin. IR treatments were by a Faxitron X-ray. DRB was used at 100 μM 2 h prior to damage.
The U2OS-DSB reporter cell line developed by the Greenberg laboratory (Tang et al. 2013) was used to monitor transcriptional repression specifically at DSBs. After 72 h of siRNA treatment, cells were left undamaged or damaged by the addition of Shield-1 (Clontech) and 1 μM 4-OHT for >3 h to induce site-specific DSBs. Both undamaged and damaged samples were treated with 1 μg/mL doxycycline to induce nascent transcription of the reporter gene. Cells were then incubated for >2 h and collected for RT-qPCR analyses using primers described in Tang et al. (2013). The transcription levels of the reporter gene from both undamaged and damaged transcription-induced samples were normalized to the undamaged transcription-induced sample for each treatment with the indicated siRNAs. This provided a comparison of the level of transcription of the reporter gene in the undamaged sample compared with the damaged sample. To analyze the recruitment of RAD51 to the FokI-mediated DSBs upon different siRNA treatments, a DSB reporter cell line without the tetracycline response elements (TREs) was used (Tang et al. 2013). DSBs were induced by adding Shield-1 and 4-OHT followed by analysis of the cells 3 h after damage using immunofluorescence with RAD51 antibodies. Nascent transcription detection by 5-ethynyl uridine (5-EU) monitoring was performed with 1 mM 5-EU added 1 h after treatment. 5-EU was detected by Click-iT RNA imaging kit (Invitrogen). Plasmid and siRNA transfections are described in the Supplemental Material.
Laser microirradiation and microscopy analysis
Laser microirradiation was carried out with a FluoView 1000 confocal microscope (Olympus) as described (Shee et al. 2013). Briefly, cells were grown on glass-bottomed dishes (Willco Wells) and presensitized with 10 μM 5-bromo-2′-deoxyuridine (BrdU) for 24 h at 37°C. A 405-nm solid-state laser was used to generate BrdU-dependent DNA damage along the laser path. Following damage, cells were analyzed by live confocal fluorescent microscopy or immunofluorescence analysis as indicated in the Supplemental Material.
Histone modification binding assays
Purified recombinant ZMYND8 (PHD–BRD) binding to modified histone peptide arrays (Active Motif) was performed. Modified histone H4 peptides were used to pull down ZMYND8 in various conditions. Details are described in the Supplemental Material.
ZMYND8 analysis by MS
ZMYND8 was purified by tandem affinity purification (TAP) and analyzed by MS as described in the Supplemental Material.
ChIP
ChIP assays were performed as previously described (Aymard et al. 2014). Briefly, AsiSI-induced DSB sites were as described before: DSB-I (Chr9) and DSB-III (Chr17) represent transcriptionally active and HR-prone regions, while DSB-1 (Chr18) and DSB-2 (Chr21) correspond to transcriptionally inactive and non-HR-prone regions. Antibodies, primers, and full details of ChIP analysis are described in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Roger Greenberg for providing the U2OS-DSB transcription reporter cell lines, Edward Marcotte for the SMARCA4 construct, and Tanya Paull for DNA-PK antibodies. We thank Blerta Xhemalce for technical support, suggestions, and insightful reading of the manuscript. We also thank Christophe Herman and Eros Lazzerini Denchi for comments on the manuscript. We are grateful to members of the Miller laboratory for their helpful discussions throughout the study. J.W.L. is funded by a Cancer Prevention Research Institute of Texas (CPRIT) post-doctoral training grant. Research in the J.S.B. laboratory is supported by the Welch Foundation (F1155) and National Institutes of Health (R21_GM099028). Research in the G.L. laboratory is supported by grants from the Association Contre le Cancer (ARC), Institut National Contre le Cancer (INCA), Agence Nationale pour la Recherche (ANR-09-JCJC-0138), Canceropole Grand Sud-Ouest (GSO), and Research Innovation Therapeutic Cancerologie (RITC). T.C. is funded by the Fondation pour la Recherche Médicale (FRM), and F.A. was funded by the ARC and a grant from the Ligue Nationale Contre le Cancer. Research in the K.M.M. laboratory is supported by start-up funds from University of Texas at Austin and CPRIT (R116). K.M.M. is a CPRIT scholar.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.252189.114.
References
- Adam S, Polo SE. 2014. Blurring the line between the DNA damage response and transcription: the importance of chromatin dynamics. Exp Cell Res 329: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam S, Polo SE, Almouzni G. 2013. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155: 94–106. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. 2013. Causes of genome instability. Annu Rev Genet 47: 1–32. [DOI] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, et al. . 2014. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21: 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. 2014. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci 111: 9169–9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett C, Krebs JE. 2011. WSTF does it all: a multifunctional protein in transcription, repair, and replication. Biochem Cell Biol 89: 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 46: 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch DB, Chiche JD, Orth D, de la Monte SM, Rosenzweig A, Bloch KD. 1999. Structural and functional heterogeneity of nuclear bodies. Mol Cell Biol 19: 4423–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM, Ferreira J, de Almeida SF. 2014. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 3: e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chen WT, Alpert A, Leiter C, Gong F, Jackson SP, Miller KM. 2013. Systematic identification of functional residues in mammalian histone H2AX. Mol Cell Biol 33: 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. 2010. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci 107: 18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciro M, Prosperini E, Quarto M, Grazini U, Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M, Christensen J, et al. . 2009. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res 69: 8491–8498. [DOI] [PubMed] [Google Scholar]
- Conomos D, Reddel RR, Pickett HA. 2014. NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat Struct Mol Biol 21: 760–770. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, Huntly BJ. 2012. Targeting epigenetic readers in cancer. N Engl J Med 367: 647–657. [DOI] [PubMed] [Google Scholar]
- Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. 2013. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell 49: 368–378. [DOI] [PubMed] [Google Scholar]
- Eichmuller S, Usener D, Dummer R, Stein A, Thiel D, Schadendorf D. 2001. Serological detection of cutaneous T-cell lymphoma-associated antigens. Proc Natl Acad Sci 98: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. 2014. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 13: 337–356. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, et al. . 2012. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149: 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, et al. . 2013. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. . 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499: 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Miller KM. 2013. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat Res 750: 23–30. [DOI] [PubMed] [Google Scholar]
- Guo R, Chen J, Mitchell DL, Johnson DG. 2011. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res 39: 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. 2008. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 8: 193–204. [DOI] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. 2010. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29: 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102: 463–473. [DOI] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. 2011. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 286: 26267–26276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha DK, Strahl BD. 2014. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat Commun 5: 3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Kunzel J, Lobrich M, Jeggo PA, Downs JA. 2014. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol Cell 55: 723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A, Oza J, Yao M, Sohail H, Ginjala V, Tomas-Loba A, Horejsi Z, Tan AR, Boulton SJ, Ganesan S. 2013. Tripartite motif-containing 33 (TRIM33) protein functions in the poly(ADP-ribose) polymerase (PARP)-dependent DNA damage response through interaction with amplified in liver cancer 1 (ALC1) protein. J Biol Chem 288: 32357–32369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, Wade PA. 2011. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 11: 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, et al. . 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 40: 976–987. [DOI] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Vermeulen W. 2012. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al. . 2010. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 190: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Park EJ, Lee HS, Lee YS, Kwon J. 2012. Genome-wide screen of human bromodomain-containing proteins identifies Cecr2 as a novel DNA damage response protein. Mol Cells 34: 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et al. . 2012. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 44: 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. . 2011. Analysis of the human endogenous coregulator complexome. Cell 145: 787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Jackson SP. 2012. Histone marks: repairing DNA breaks within the context of chromatin. Biochem Soc Trans 40: 370–376. [DOI] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17: 1144–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Kim H, Laurent B, Zhao Y, Lim HJ, Majid MC, Dango S, Luo Y, Hempel K, Sowa ME, et al. . 2013. The histone demethylase LSD1/KDM1A promotes the DNA damage response. J Cell Biol 203: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30: 2135–2146. [DOI] [PubMed] [Google Scholar]
- Pai CC, Deegan RS, Subramanian L, Gal C, Sarkar S, Blaikley EJ, Walker C, Hulme L, Bernhard E, Codlin S, et al. . 2014. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat Commun 5: 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. 2012. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem 287: 6764–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Micci F, Thorsen J, Haugom L, Buechner J, Kerndrup G, Tierens A, Zeller B, Heim S. 2013. Fusion of ZMYND8 and RELA genes in acute erythroid leukemia. PLoS ONE 8: e63663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Bonhomme C, Chen D, Soutoglou E. 2012. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol 19: 276–282. [DOI] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. 2006. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J 25: 3986–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter AC, et al. . 2014. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Reports 7: 2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes & Dev 25: 409–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. 2010. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J 29: 3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. 2013. Chromatin remodeling at DNA double-strand breaks. Cell 152: 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al. . 2011. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145: 692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al. . 2010. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 30: 3582–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C, Cox BD, Gu F, Luengas EM, Joshi MC, Chiu LY, Magnan D, Halliday JA, Frisch RL, Gibson JL, et al. . 2013. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. eLife 2: e01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847. [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. 2010. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190: 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G. 2012. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46: 722–734. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. 2009. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 11: 1376–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ. 2010. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci 35: 333–338. [DOI] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. . 2010. TRIM24 links a non-canonical histone signature to breast cancer. Nature 468: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Matsuura M, Sugawara M, Ushijima M, Miyata S, Nagasaki K, Noda T, Miki Y. 2014. Development of detection method for novel fusion gene using GeneChip exon array. J Clin Bioinforma 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al. . 2014. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. 2011. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 11: 481–492. [DOI] [PubMed] [Google Scholar]
- Zeng W, Kong Q, Li C, Mao B. 2010. Xenopus RCOR2 (REST corepressor 2) interacts with ZMYND8, which is involved in neural differentiation. Biochem Biophys Res Commun 394: 1024–1029. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.