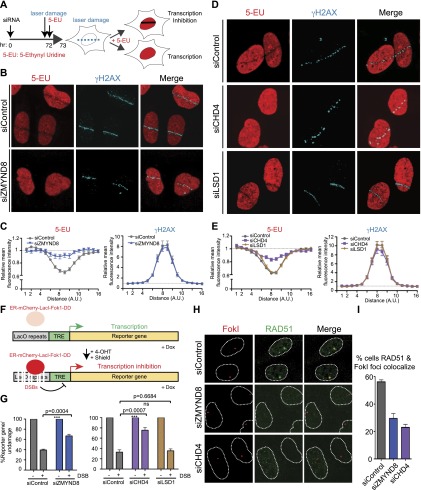
Figure 6.
ZMYND8 and CHD4 mediate transcriptional repression upon DNA damage. (A) Scheme of nascent transcription analysis by 5-ethynyl uridine (5-EU) monitoring following laser damage. (B) ZMYND8 promotes transcriptional repression following laser damage. Cells treated with control or ZMYND8 siRNAs were subjected to the scheme shown in A and analyzed by immunofluorescence. γH2AX marks DNA damage. (C) Quantification of 5-EU and γH2AX fluorescence intensity from B. Measurements of fluorescent intensity along lines perpendicular to the laser damage, which contained both damaged and undamaged regions, were obtained. Values were normalized to undamaged regions. Error bars indicate SEM; n > 10. (D) CHD4 but not LSD1 is required for transcriptional repression following laser damage. Experiments performed as in B. (E) Quantification of D as in C. (F) Scheme of U2OS DSB reporter cells adapted by permission from Macmillan Publishers Ltd. from Tang et al. (2013), © 2013. Shield-1 and 4-OHT regulate the Fok1 nuclease, which induces DSBs upstream of reporter genes within LacO repeats. Doxycycline induces transcription of the reporter gene, allowing transcriptional repression upon DSB induction to be measured by quantitative PCR (qPCR). (G) ZMYND8 and CHD4, but not LSD1, regulate transcriptional repression at DSBs. The system from F was analyzed in cells treated with the indicated siRNAs. Error bars indicate SEM; n = 4. P-values were calculated using Student’s t-test. (H) ZMYND8 and CHD4 promote RAD51 loading at DSBs. RAD51 loading by immunofluorescence in siControl, siZMYND8, and siCHD4 cells was analyzed 3 h after DSB induction using a FokI-inducible DSB system (Tang et al. 2013). (I) Quantification of H. Error bars indicate SEM; n = 3. siRNA #1 was used for siZMYND8 treatments.