Fig. 1. RNAi Vectors for HF Therapy.
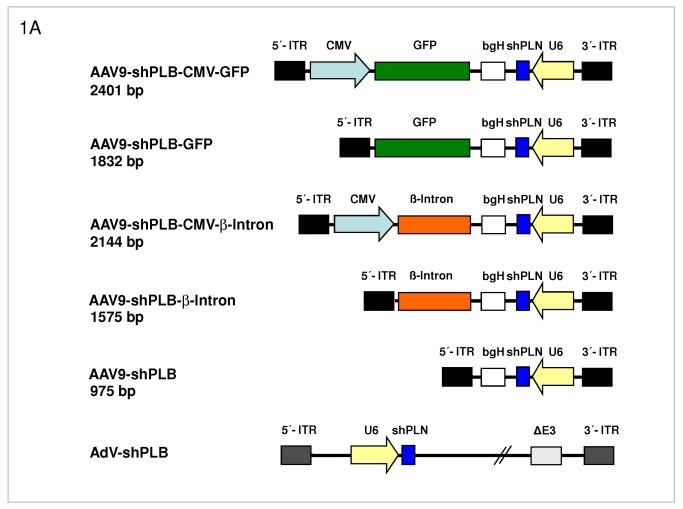
A: Maps of the RNAi vector genomes. Self-complementary “dimeric” AAV genomes (rAAV2) were pseudotyped into rAAV6 or rAAV9 capsids for studies in cardiomyocytes in vitro or HF rats in vivo, respectively. rAAV-shPLB has the same U6-shRNA transcription system as AdV-shPLB and the corresponding shGFP control vectors. Two further rAAV vectors carried CMV-GFP or CMV-β-intron expression cassettes in head-to-head orientation with the U6-shPLB sequence and separated from them by a bovine growth hormone (bgH) terminal signal. To assess the influence of the CMV promoter on vector function the CMV-free variants rAAV-shPLB-GFP and rAAV-shPLB-β-intron were constructed. Please note that the genomes in the rAAV6 vectors for in vitro work are identical to those shown for the rAAV9 vectors shown here.
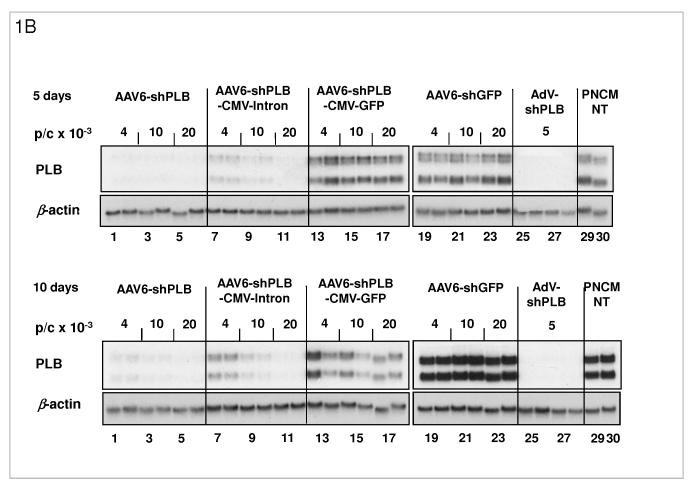
B: Comparison of the target silencing efficacy of shRNA vectors in NRCMs. Cells were harvested 5 (upper part) or 10 days (lower part), respectively, after treatment with the respective vector at the dose in particles per cell (p/c) given above the lanes. Northern blots were then carried out using a rat PLB-specific probe. To confirm equal RNA loading the blots were striped and rehybridized with a β-actin-specific probe. Lanes 1 to 18 show dose dependency of RNAi-mediated PLB-mRNA downregulation for the rAAV-based vectors rAAV-shPLB (lane 1-6), rAAV-shPLB-CMV- β-intron (lane 7-12), and rAAV-shPLB-CMV-GFP (lane 13-18). Lanes 19-24 show as a control for unspecific shRNA effects PLB-mRNA expression after treatment with rAAV-shGFP which generates an shRNA sequence targeting GFP (lanes 19-24). There was no difference towards untreated cells (lanes 29,30). For comparison with rAAV the adenovector AdV-shPLB (lanes 25-28) was used. PLB-mRNA was ≥98% ablated until day 10 by rAAV-shPLB at the lowest dose of 4×103 p/c (lanes 1,2), similar to AdV-shPLB (lanes 25-28). Incorporation of a CMV-GFP expression cassette in the rAAV-shPLB vector (lanes 7-12) to provide this vector with a tag which is easily detectable by in vivo imaging, led to strong GFP expression in infected cells (not shown) but unexpectedly abolished its PLB gene silencing effect. Incorporation of a CMV-β-intron cassette (lanes 12-18) had a similar but less pronounced effect. We therefore used only rAAV9-shPLB vs rAAV9-shGFP and AdV-shPLB vs AdV-shGFP for in vivo therapy (Fig. 2a-g and 3a-f).
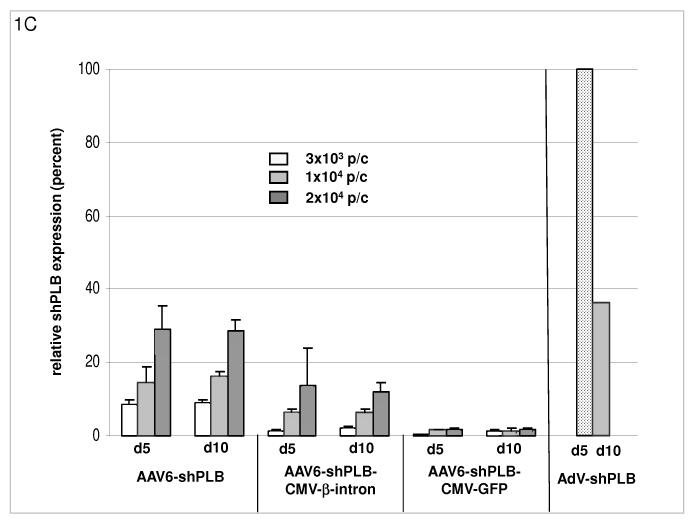
C: The cellular shRNA levels produced by the vectors from panel B. In the presence of a CMV-GFP cassette shPLB production was abolished (lanes 13-18), whereas the U6-shPLB vector without additional sequences showed stable expression over 5 (lanes 1-3) and 10 days (lanes 4-6). AdV-shPLB generated very high shPLB levels on day 5 which then declined rather rapidly in NRCMs. Studies with further vectors (Suppl. Fig. 1a-c) showed that interaction of the CMV promotor with the shRNA-transcribing polymerase type III U6 promotor apparently disturbs shRNA transcription from the respective AAV vectors.
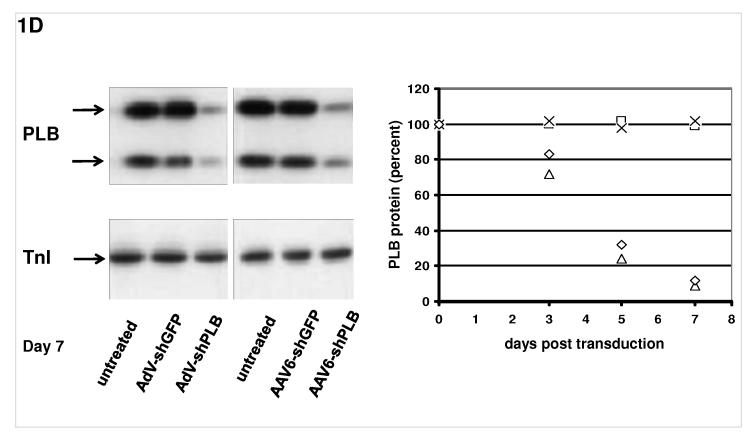
D: On the left a Western blot analysis of PLB protein during treatment of NRCMs and on the right its quantitation on days 3, 5, and 7 after vector addition is shown. AdV-shPLB and rAAV6-shPLB resulted on day 7 in downregulation of cellular PLB to 9 % and 13 %, respectively, of baseline.
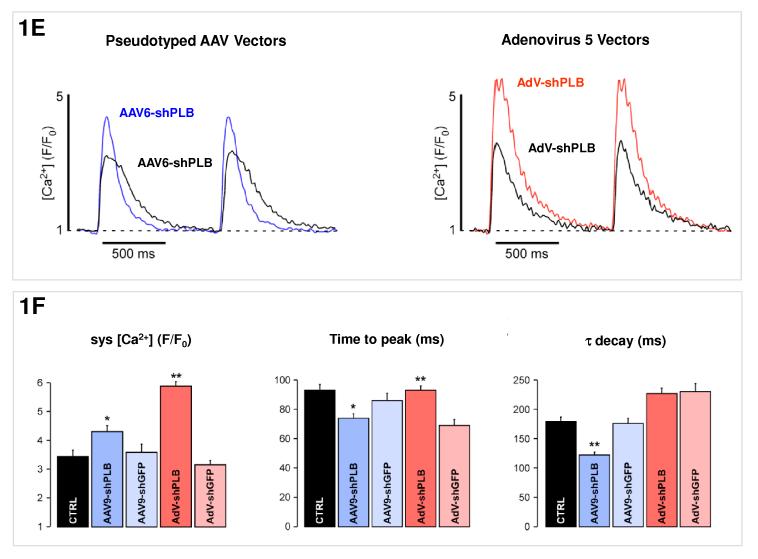
E: [Ca2+]i transients in NRCMs during AAV9-shPLB treatment showed significantly higher amplitudes and accelerated transient kinetics (shortened TTP and τ) compared to the AAV9-shGFP group with transients indistinguishable from untreated cells. AdV-shPLB treatment also resulted in a significantly higher amplitude than in AdV-shGFP controls. In contrast to the AAV9 groups, TTP was prolonged in the AdV-shPLB vs. AdV-shGFP group which displayed no difference in τ.
F: Statistical evaluation of the [Ca2+]i transients. * denotes p<0.05 and ** p<0.01. Additional studies of sarcoplasmic reticulum (SR) Ca2+ loading in the adenovirus groups (measured by rapid caffeine addition) were performed (Suppl. Fig. 1d,e) and showed increased SR Ca2+ loading and fractional Ca2+ release from the SR in AdV-shPLB vs. AdV-shGFP group.