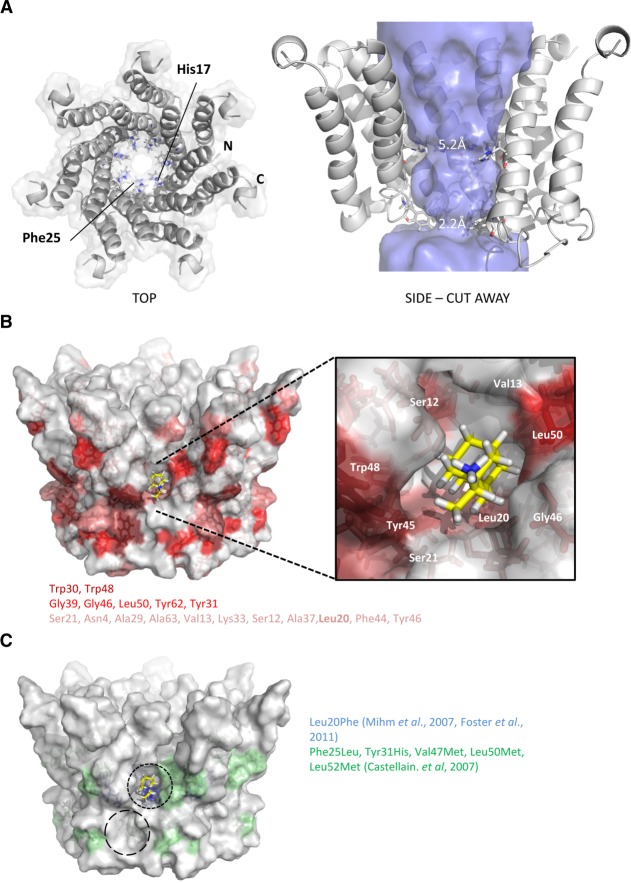
Figure 3.
Construction of heptameric channel models based on monomeric p7 NMR structure. Energy minimized channel models were constructed in Maestro (see Materials and Methods) by symmetrical arrangement of individual biological unit protomers with multiple rounds of energy minimization. Stable structures resulted from a tilted orientation of individual protomers relative to the predicted membrane normal. The N-terminal helices formed the channel lumen and the C-terminal helices stabilized the channel complex, consistent with previous models and predictions. (A) Left: top-down surface view showing individual protomers as ribbons. His17 and Phe25 are highlighted projecting into the lumen. Right: side view cutaway showing orientation of individual protomers, His17 and Phe25 highlighted and lumenal diameter indicated by the “Hollow” program.31 (B) Residue positions in context of heptameric channel model seen to interact in NMR experiments (color grading as in Fig. 2C). (C) Amino acid changes in patients nonresponsive to amantadine in two separate studies as indicated. Predicted rimantadine docking is shown for one of the seven primary peripheral binding pockets (black dashed line) along with a potential secondary binding site (longer dashed circle).