Abstract
A crucial step in the development of therapeutic monoclonal antibodies is the selection of robust pharmaceutical candidates and screening of efficacious protein formulations to increase the resistance toward physicochemical degradation and aggregation during processing and storage. Here, we introduce small-angle X-ray scattering (SAXS) to characterize antibody solution behavior, which strongly complements conventional biophysical analysis. First, we apply a variety of conventional biophysical techniques for the evaluation of structural, conformational, and colloidal stability and report a systematic comparison between designed humanized IgG1, IgG2, and IgG4 with identical variable regions. Then, the high information content of SAXS data enables sensitive detection of structural differences between three IgG subclasses at neutral pH and rapid formation of dimers of IgG2 and IgG4 at low pH. We reveal subclass-specific variation in intermolecular repulsion already at low and medium protein concentrations, which explains the observed improved stability of IgG1 with respect to aggregation. We show how excipients dramatically influence such repulsive effects, hence demonstrating the potential application of extensive SAXS screening in antibody selection, eventual engineering, and formulation development.
Keywords: IgG antibody, stability, analysis, protein formulation, protein aggregation, physicochemical properties, small-angle X-ray scattering (SAXS)
INTRODUCTION
Immunoglobulin G (IgG) monoclonal antibodies are one of the largest classes of biopharmaceuticals because of their high antigen specificity and long half-lives in the body. IgG is the most abundant antibody isotype in blood and external tissue, where it controls infections of the body.1 The four human IgG subclasses IgG1, IgG2, IgG3, and IgG4 account for about 60%, 25%, 10%, and 5% of IgG concentration in blood, respectively.2 Today, more than 20 antibodies are approved for clinical use in Europe or the United States, and more than 200 are in clinical development. A majority of the approved antibodies are of the IgG1 subclass, whereas there is a greater variation of IgG subclass for those in clinical development. Several antibodies have also been engineered to modify known effector functions.3
In the process of selecting and/or developing therapeutic IgG monoclonal antibodies, it is important to evaluate the biological function and physicochemical characteristics of the IgG formats. The monoclonal antibodies are susceptible to deterioration during manufacture and storage, which could lead to unwanted immune response and decreased bioactivity.4,5 Therefore, it is crucial to assess the physical and chemical stability of antibody proteins including aggregation, fragmentation, deamidation, and so on at different formulation conditions. Several techniques have been developed for physicochemical characterization, and applied in screening for selection and development of therapeutic monoclonal antibody (mAb) as well as for formulation screening. However, limitations of the individual techniques are recognized. As an example, size-exclusion HPLC (SE-HPLC) is routinely used in pharmaceutical industry for characterizing aggregation and fragmentation. However, buffer exchange with the mobile phase of SE-HPLC can in some cases alter the solution behavior of the antibodies. Nonspecific interactions between the protein sample and the column matrix can also deleteriously affect the results and the conformation of the protein.6 In addition, another widely used technique, dynamic light scattering (DLS), exhibits superiority for high-throughput screening in terms of detecting aggregation. However, the resolution is relatively low with respect to separation of different particle populations and the influence of excipients on the apparent hydrodynamic radius (Rh) has to be addressed.
Small-angle X-ray scattering (SAXS) has been widely used for the analysis of biological macromolecules in solution7 and can monitor the changes of protein conformation,8 transient protein–protein interactions,9 intermolecular attraction and repulsion,10 oligomerization, and aggregation.11 Thus, the most unique feature of SAXS data analysis is the inclusion of information about both conformational and colloidal stability. Here, we introduce SAXS analysis of the solutes in a setup that is compatible with high-throughput screening, and compare the readily accessible information content from SAXS data with a number of well-established methods. We present a systematic study of humanized IgG1, IgG2, and IgG4 with identical variable regions. These proteins have been produced recombinantly and prepared to a high quality with a purity of more than 99%, enabling direct comparison of the three IgG subclasses. Several methods are applied under the same experimental conditions while using the same protein batch for all analyses. In comparison with conventional techniques, we outline the advantages of SAXS analysis and envision the useful application of high-throughput SAXS screening as part of an efficient and multifaceted strategy on selection and development as well as preformulation development of therapeutic antibodies.
MATERIALS AND METHODS
Materials
The complementarity-determining regions (CDRs) from anti-TNP (2,4,6-trinitrophenol) mouse mAb were grafted into human variable regions,12,13 which were then joined to the constant regions of human IgG4. The IgG1 and IgG2 were constructed by replacing the constant region of IgG4 heavy chain, thus the three IgGs have identical light chains and variable regions of the heavy chain. A single mutation S228P (Kabat numbering14) was included in the IgG4 heavy chain to prevent half mAb generation. Double gene constructs were created by cloning the light chain and heavy chain cDNA into the mammalian expression vector pEE14.4 (Lonza Biologics, Auckland, New Zealand). The stable cell lines were generated by transfecting the recombinant plasmids into CHOK1SV cells (Lonza Biologics). The antibodies were purified from cell culture supernatants using protein A-based methods and gel filtration chromatography with identical purification protocols. In order to monitor the glycosylation patterns, the antibodies were digested by PNGase F, then the released glycans were labeled with 2-aminobenzamide and analyzed by ACQUITY UPLC® System (Waters, Milford, Massachusetts) with BEH Glycan Separation Technology Column (GST column) coupled to the fluorescence detector. The purified samples were then formulated at 15–25 mg/mL in phosphate-buffered saline (PBS) at pH 6.9 and sterile filtered using 0.22 μm polyvinylidene difluoride low-binding-syringe-driven filter units from Millipore (Carrigtwohill, Ireland). Concentrations were determined based on A280 using theoretical extinction coefficients 14.81 (IgG1), 14.52 (IgG2), and 14.85 (IgG4) (g/100 mL)−1 cm−1, respectively. The bulk solution was flash frozen in liquid nitrogen and stored at −80°C.
Sample preparations particular for each method are described in relevant sections, and the experimental conditions, including formulation buffers and storage conditions, are summarized in Table1.
Table 1.
Summary of the Formulation Conditions
| Base Buffera | pHb | Excipientc | Storage Conditions | Methods |
|---|---|---|---|---|
| Initial Screening of pH and Additives | ||||
| Na-citrate | 3.0 | 100 mM NaCl, 0.25 M sucrose, 0.01% Tween 80 | Initial/no storage; 40°C, 4 days; 2 mg/mLd | DLS, DSF |
| Glycine | 3.5 | |||
| Na-acetate | 4.0 | |||
| Na-acetate | 4.5 | |||
| Na-acetate | 5.0 | |||
| Succinate | 5.5 | |||
| Histidine | 6.0 | |||
| Histidine | 6.5 | |||
| Imidazole | 7.0 | |||
| Glycyl–glycine | 8.0 | |||
| Glycine | 9.0 | |||
| Glycine | 10.0 | |||
| Stability Studies Under Accelerated Storage Conditions | ||||
| Na-citrate | 3.3 | 100 mM NaCl, 0.25 M sucrose, 0.05% Tween 80 | Initial/no storage; 40°C, 8 weeks; 12 mg/mLd | SAXSe, UV280 nm, SE-HPLC, LabChip |
| Na-acetate | 5.0 | |||
| Histidine | 6.5 | |||
| Na-phosphate | 7.4 | |||
| Tris–HCl | 8.5 | |||
| Stability Studies Under Normal Storage Conditions | ||||
| Histidinef | 6.5 | 0.25 M sucrose | Initial/no storage; 5°C, 8 weeks; 25°C, 8 weeks; 2 mg/mLd | SE-HPLC, iCE |
| Na-phosphateg | 7.4 | 100 mM NaCl | ||
Final concentration of the base buffers was 50 mM.
Measured at room temperature.
Excipients were included respectively to each of the base buffers.
Antibody concentration during storage.
Only the samples at initial time point were measured by SAXS.
Formulation A.
Formulation B.
Analytical Methods
In this study, we combine conventional biophysical analysis with high-throughput compatible SAXS-based screening. For the conventional analysis, we performed (1) an initial screening of pH and excipients using differential scanning fluorescence (DSF) and DLS (after storage at 40°C for 4 days) in a high-throughput setup; (2) the physical and chemical stability of a selection of samples (Table1) was further studied by UV280 nm, SE-HPLC, and automated microfluidic SDS electrophoresis (LabChip) under accelerated storage conditions (40°C); and (3) two formulations were selected for further studies under normal storage conditions (5°C and 25°C) and characterized by DLS, SE-HPLC, LabChip, and imaged capillary isoelectric focusing (iCE). In addition to these conventional analyses, the solution behavior of antibodies was investigated by SAXS, prior to any heat stressing and long-term storage. LabChip and iCE are described in the Supplementary Material.
Differential Scanning Fluorescence
The antibodies were buffer exchanged using illustra™ NAP™-5 Columns (GE healthcare, Uppsala, Sweden) into 5 mM histidine, pH 6.5, and the concentration was adjusted to 4 mg/mL using the same buffer. Before the DSF analysis, the initial 4 mg/mL protein stock was diluted with water to 0.6 mg/mL and then mixed with an equal volume of buffer stock containing 5X SYPRO® orange dye, which was supplied as 5000X-concentrated solutions in dimethyl sulfoxide (Invitrogen, Eugene, Oregon). The final volume of reaction mixtures was 50 μL in each well. Ninety-six-well optical reaction plates were used (VWR International, Radnor, Pennsylvania). The fluorescence signal was detected using a MyiQ single-color real-time PCR detection system (Bio-Rad Labs, Berkeley, California). A 480-nm excitation filter with 40-nm bandwidth and a 540-nm emission filter with 50-nm bandwidth were used. Temperature scans were performed from 25°C to 90°C at a scan rate of 1°C/min, and thermal transition midpoint (Tm) was determined using in-house software developed by Novo Nordisk. The unfolding of an IgG is complex and the individual domains of the conserved and variable regions unfold independently. For simplicity, we only present data for the first unfolding transition of each IgG. This transition represents the unfolding of the CH2 domain in the fragment crystallizable (Fc) region.15
Dynamic Light Scattering
Samples were prepared in the same manner as for the DSF analysis, with the difference that before the DLS analysis the final sample was prepared by mixing 10 μL of protein stock (4 mg/mL) with 10 μL of a buffer stock (Table1). All samples were analyzed at the initial time point and after storage at 40°C for 4 days. The DLS measurements were carried out in a 384-well microplate at 25°C using a Dynapro plate reader (Wyatt Technology, Goleta, California) equipped with an 831-nm laser. A total of 10 acquisitions of 5 s each were collected with auto adjustment of laser power; autoattenuation was applied. Solvent properties were edited for each sample to adjust the viscosity and refractive index. All formulation conditions were studied in duplicate wells. We manually checked the data quality of each sample according to the fit between correlation curve and exponential decay function. The Rh, normalized intensity, and polydispersity were calculated from the correlation function using DYNAMICS 7 (Wyatt Technology).
Size-Exclusion HPLC
Protein samples in a wide range of different formulation buffers (Table1) were concentrated to approximately 12 mg/mL using 30,000 MWCO Amicon Ultra-4® centrifugal filters (Millipore). The samples were stored at 40°C. After 8 weeks incubation, the samples were centrifuged at 13,523 g for 10 min to remove precipitates. The remaining protein concentration was determined based on A280. The samples representing normal storage conditions at 5°C and 25°C were investigated in two formulations: 20 mM histidine (pH 6.5) containing 0.25 M sucrose (buffer A) and 50 mM sodium phosphate (pH 7.4) containing 100 mM NaCl (buffer B). The samples were exchanged into the formulation buffers by using illustra™ NAP™-5 Columns (GE Healthcare), and their concentrations were adjusted to 2 mg/mL. These samples were stored at 5°C and 25°C, respectively, and analyzed after 4 and 8 weeks, respectively. The amounts of aggregates (high-molecular-weight species) and degradation species (low-molecular-weight species, LMWS) were analyzed by SE-HPLC using a G3000SWXL column (Tosoh Corporation, Tokyo, Japan). The mobile phase consisted of PBS (Invitrogen, Paisley, UK) and the flow rate was 0.8 mL/min. Twenty microgram of antibody was injected in a volume of 10 μL. The UV absorbance peaks were detected at 215 and 280 nm.
Small-Angle X-ray Scattering
The IgG antibodies were analyzed by SAXS without heat stressing or long-term storage. The formulation setup is the same as SE-HPLC analysis under accelerated storage conditions (Table1). Before measurements, samples were diluted into concentration series, following beamline standard procedures.16 Samples were centrifuged in a fixed angle rotor centrifuge at 15,871 g for 10 min immediately prior to measurement. The synchrotron SAXS data were collected on beamline X33 at the European Molecular Biology Laboratory on the DORIS III storage ring (DESY, Hamburg, Germany). Samples were loaded using the automated sample changer. Scattering from the IgG protein solutions with concentrations between 1 and 12 mg/mL was measured at 8°C in the momentum transfer ranges of 0.07–5.0 nm−1 [s = 4πsin(θ)/λ, where 2θ is the scattering angle and λ is the X-ray wavelength (λ = 1.5 Å)]. Data analysis was performed using the software suite ATSAS.16 In order to eliminate the impact of structure factors,10 the low-concentration data of each sample was merged with high-concentration data after superposition of the curves in areas where scattering patterns were identical for all concentrations. All SAXS curves were scaled according to the curve of the same antibody in Na-phosphate (pH 7.4) buffer with NaCl. The radius of gyration (Rg) and the scattering intensity at zero angle I(0), for each sample was determined from the Guinier approximation. The pair distance distribution functions, P(r), were evaluated using GNOM.17
RESULTS
Three humanized IgG subclasses, IgG1, IgG2, and IgG4, were designed and expressed with identical anti-TNP CDRs, and purified to >99% purity, according to SE-HPLC profiles (Fig.1a, nonstressed samples). Glycan analysis (see Supplementary Material) further revealed that the glycosylation patterns of the three recombinant batches are essentially identical. This experimental design enables an extensive systematic comparative analysis of a number of solution conformation and stability parameters from the three different IgG subclasses. We show how SAXS screening, applying robotics for the sample handling18 and semiautomated primary data analysis,19,20 readily provides valuable information about the antibody solution behavior, which strongly complements the information available from conventional analytical methods.
Figure 1.
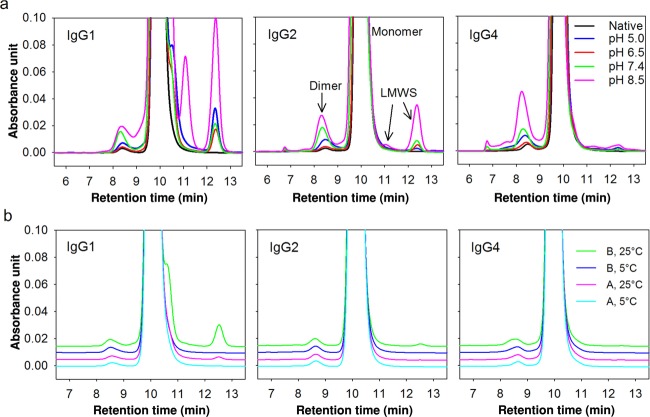
(a) Stability of antibodies investigated by SE-HPLC under accelerated storage conditions (40°C for 8 weeks). LMWS indicate the low-molecular-weight species. Blue, red, green, and pink trace lines indicate the samples at pH 5.0, 6.5, 7.4, and 8.5, respectively, containing 100 mM NaCl. The black trace line indicates the nonstressed sample at pH 7.4. (b) Stability of antibodies investigated by SE-HPLC under normal storage conditions (5°C or 25°C for 8 weeks). Trace lines indicate the samples in Formulation A at 5°C (cyan), Formulation A at 25°C (pink), Formulation B at 5°C (blue), and Formulation B at 25°C (green). Formulation A: 50 mM histidine, pH 6.5, 250 mM sucrose. Formulation B: 50 mM Na-phosphate, pH 7.4, 100 mM NaCl.
Characterization of Antibody Stability by Conventional Analytical Methods
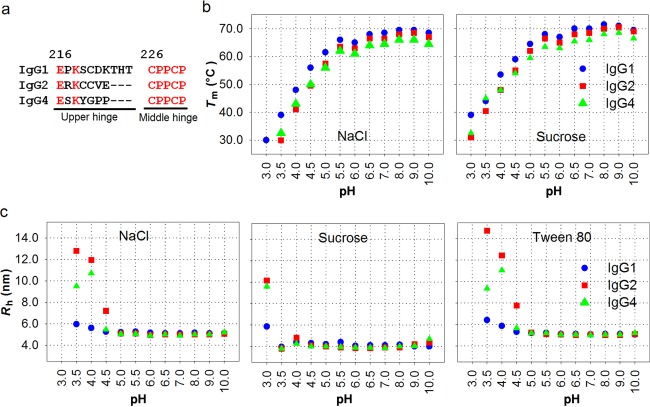
A comparison of the conformational stability of IgG1, IgG2, and IgG4 was performed using thermally induced unfolding experiments. The antibodies were investigated in the pH region 3–10, monitored with DSF. As shown in Figure 2b, the thermal denaturation of the antibodies exhibited pH-dependent profiles and the thermal stability increased dramatically from pH 3.0 to 5.5 for all three antibodies. The first transitions of IgG2 and IgG4 at pH 3.0 containing NaCl were not seen on the DSF curves, indicating that the CH2 domains of these two subclasses unfold at temperatures lower than 25°C at this pH value. Because of the nonspecific interactions of Tween 80 with the Sypro Orange dye, no DSF data are available for Tween 80 formulations (data not shown). The fluctuation at pH 6.0 on each line was because of the influence of buffer species as previously reported.21,22 IgG1 showed the highest Tm values and hence exhibits the highest thermal stability for the whole pH region tested, whereas the presence of sucrose significantly improved the thermal stability for all three subclasses. Both of these features are especially evident at the lower pH values.
Figure 2.
(a) Primary structure of IgG1, IgG2, and IgG4 hinge regions (Kabat numbering14). (b) Tm measured by DSF and (c) Rh measured by DLS at various pH values and by inclusion of various excipients. IgG1 (blue), IgG2 (red), and IgG4 (green). DLS measurements were performed on stressed samples that were stored at 40°C for 4 days.
Using the exact same experimental conditions as for the DSF analysis, the aggregation behavior was monitored by measuring the Rh of the samples at the initial time point (data not shown) and after storage at 40°C for 4 days (Fig.2c). In accordance with the observed effect of pH on the DSF analysis, increasing aggregation was observed at lower pH, whereas no changes were observed after storage above pH 5.0. Below pH 5.0, IgG1 showed greatest resistance to aggregation and its Rh value was only significantly increased at pH 3.0, hence suggesting a potential link between conformational stability and aggregation during storage. IgG2 showed higher Rh value than IgG4 between pH 3.5 and 4.5. Sucrose significantly decreased aggregation of all IgG subclasses at low pH, which can be explained by the theory of preferential hydration.23–25 In our studies, the nonionic surfactant Tween did not significantly reduce aggregation.
Accelerated studies, probing the physical and chemical stability of the antibodies under a number of experimental conditions (Table1), were assessed by UV280 nm, SE-HPLC, and LabChip (see Supplementary Material). The samples were heat-stressed at 40°C for 8 weeks. SE-HPLC quantitatively measures the aggregates, dimers, and fragments of antibodies, whereas LabChip was employed to investigate the fragmentation within the domains and the nonreducible chemical cross-linking. Because of the aggregation and precipitation, the SE-HPLC and LabChip profiles of the samples at pH 3.3 are not available (Supplementary Fig. S3a). As shown in Figure 1a, the SE-HPLC elution profiles of the other samples clearly revealed the presence of soluble aggregates, dimers, monomers, and fragments. Significant differences between subclasses were observed. The amount of soluble aggregates and dimers of IgG4 increased more than for IgG1 and IgG2 at pH 8.5, as well as in the presence of Tween 80 (Supplementary Fig. S3b). Aggregation levels of all IgG subclasses exhibited pH dependence with the highest stability in the histidine buffer at pH 6.5, which is indeed frequently chosen when developing protein formulation in the pharmaceutical industry.21,26 In the entire pH range studied, more fragments were formed by IgG1 (Fig.1a), resulting in the fragmentation profiles IgG1 > IgG2 > IgG4 at the same formulation conditions (i.e., IgG4 being the most stable). Fragmentation did not significantly differ among additives tested (Supplementary Fig. S3b). The fragmentation of antibodies observed at pH 5.0, 6.5, and 7.4 was qualitatively similar to those reported by others.26,27 However, the larger pH range studied here revealed the most significant differences in fragmentation at pH 8.5.
Antibody storage stability after 8 weeks at 5°C and 25°C was further studied in two buffers. On the basis of the DLS and SE-HPLC results in accelerated stability studies at 40°C, aggregation is decreased in the presence of sucrose (Fig.2c), whereas fragmentation is lowered in histidine buffer (Fig.1a), hence 20 mM histidine at pH 6.5 containing 0.25 M sucrose is selected as an optimized formulation (Formulation buffer A). Na-phosphate (50 mM) pH 7.4 containing 100 mM NaCl (Formulation buffer B), which is commonly used in biological research, is selected for comparison. No aggregation or fragmentation was observed by SE-HPLC (Fig.1b) and LabChip analysis (Supplementary Information) for any of the samples stored at 5°C (Fig.1b). At 25°C, insignificant amounts of fragments for IgG2 and substantial amounts for IgG1 were observed (Fig.1b). According to the fragmentation profiles, the antibodies are more stable in Formulation A and IgG4 exhibited the best stability. Combined with results from accelerated studies, all three IgG subclasses exhibit similar profiles in terms of aggregation at pH 6.5 (Figs. 1a and 1b), thus antibody fragmentation becomes the dominant consideration when comparing IgG subclasses at this pH. Considering that larger particles might be lost when measuring by SE-HPLC, DLS was also employed to investigate aggregation. No differences were observed in the Rh for any of the samples (data not shown).
In conclusion, the physical stability in terms of aggregation can be ranked IgG1>IgG4>IgG2 below pH 5.0, whereas IgG1 > IgG2 > IgG4 above pH 5.0. The chemical stability in terms of fragmentation can be ranked IgG4 > IgG2 > IgG1 at all of the formulation conditions investigated (pH, additives, and temperatures).
Solution Behavior of Antibodies Investigated by SAXS
According to thermal stability analysis by conventional biophysical methods, the antibodies exhibit significant differences in terms of aggregation amongst pHs, additives, and subclasses. In order to further investigate the solution behavior of antibodies at the initial time point, SAXS was employed. We used the same formulation setup as for SE-HPLC analysis in accelerated stability studies (Table1) but without heat stressing or long-term storage. Considering that the antibodies are prone to aggregate at low pH, the low pH samples were measured immediately after exchanging the buffer into 50 mM Na-citrate (pH 3.3).
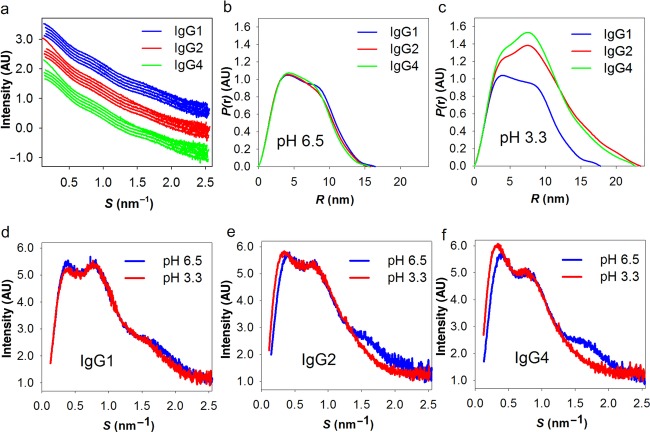
The experimental SAXS curves from the samples containing NaCl are shown in Figure 3a. From the indirect Fourier transformation of the scattering data,17 the P(r) of the antibodies in solution is obtained (Fig.3b), and from the slightly different shapes it is evident that subtle differences exist in the overall solution conformation between different subclasses. From the P(r), the maximal dimension (Dmax) of the solute species is also estimated. Dmax is comparable for all three subclasses, yet the larger dimensions are on average reached more often by the IgG1 molecules [as seen by a small right-shift of the P(r) curve for IgG1]. Indeed, IgG1 has a longer hinge region than IgG2 and IgG4 (Fig.2a), and thus may either have a more extended conformation or greater overall flexibility in solution. Accordingly, IgG1 reveals slightly higher Rg values than the other IgGs (with the exception of the partially dimerized samples at low pH, Fig.4a). The differences between these curves are all reflections of the IgG subclass variation with respect to solution conformation(s). These differences can be investigated further and potentially modeled, but this is beyond the scope of this publication.
Figure 3.
(a) Comparison of the SAXS curves at different pH. The SAXS curves of the samples at pH 3.3, pH 5.0, pH 6.5, pH 7.4, and pH 8.5 are translated for comparison and ordered sequentially from top to bottom for each IgG (i.e., within each color). (b and c) The P(r) of the samples at pH 6.5 and pH 3.3 from the indirect Fourier transformation of the scattering intensity. (d–f) Kratky plots based on the SAXS data of IgG1, IgG2, IgG4, respectively, at pH 6.5 and 3.3.
Figure 4.
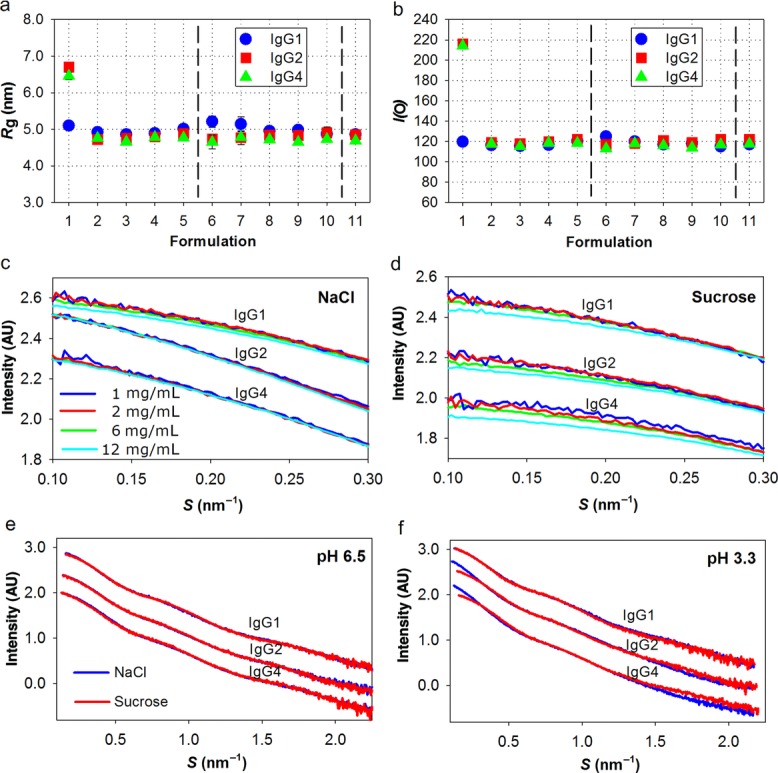
(a) Rg of three IgG subclasses in different formulation buffers and (b) the corresponding I(0) values. 1–5 on x-axis indicate the samples at pH 3.3, 5.0, 6.5, 7.4, and 8.5, respectively, containing 100 mM NaCl. 6–10 indicate the samples at pH 3.3, 5.0, 6.5, 7.4, and 8.5, respectively, containing 250 mM sucrose. 11 indicate the samples at pH 7.4 containing 0.05% Tween 80. (c and d) Concentration dependence of the three antibodies in Na-citrate buffer (pH 3.3). The data curves of the individual IgGs have been transposed for clarity. (e and f) SAXS curves comparing the effect of sucrose and NaCl at pH 6.5 and 3.3. Dimerization is observed at pH 3.3 for IgG2 and IgG4. The data curves of the individual IgGs have been transposed for clarity.
However, within each antibody subclass, all SAXS curves are similar from pH 5.0 to pH 8.5 (Fig.3a), which indicates that each antibody has the same overall conformation in this pH region. In contrast, at pH 3.3, both IgG2 and IgG4 exhibit significantly different SAXS curves and subclass differences are also more evident (Fig.3c). Including also the comparison of Kratky plots at pH 6.5 and 3.3 for each of the three subclasses (Figs. 3d–3f), it is evident that larger scatterers appear in the IgG2 and IgG4 low pH solutions. The overall size of these larger scatterers are well-resolved within the resolution range (a peak at low angles is clearly resolved), which shows that samples are devoid of large (unspecific) aggregates. In contrast, in the presence of large aggregates, the curves would have revealed a pronounced increase in scattering intensity in this region. The Rg calculated from the Guinier region of the SAXS curve, shows the increased sizes in IgG2 and IgG4 (Fig.4a). In addition, the extrapolated I(0) is proportional to the molecular mass of the protein,28 thus (partial) oligomerization or aggregation will increase I(0). At almost all experimental conditions investigated, the calculated MW is as expected for the monomeric antibody with only one remarkable difference: the I(0) values of IgG2 and IgG4 at pH 3.3 with NaCl were nearly doubled compared with measurements at other pH values (Fig.4b). Also, there are no notable effects on I(0), and thus the average oligomeric state, when varying the total protein concentration (Figs. 4c and 4d). In conclusion, the increased MW of IgG2 and IgG4 at pH 3.3 is because of the specific dimerization at early time point after buffer exchange. Within the time-range observed, there are no signs of further oligomerization or aggregation in any of the samples investigated.
The SAXS curves of the samples at pH 3.3 and pH 6.5 with different additives are compared in Figures 4e and 4f. IgG2 and IgG4 dimerization at pH 3.3 was significantly decreased by the addition of sucrose. Likely, this is because of the preferential exclusion effects.29 Mosbaek et al.10 reported that repulsion was the dominating interparticle effect in IgG2 samples with sucrose at protein concentrations below 38 mg/mL. According to the concentration series at pH 3.3 measured in this analysis (Figs. 4c and 4d), similar repulsion was observed for all IgG samples with sucrose. The repulsion reduces the proximity of the molecules and thus improves the colloidal stability of the antibodies. The SAXS curves for all subclasses at pH 6.5 exhibit only minor differences between sucrose and NaCl formulations, which can be systematically modeled by a constant scattering contribution derived from solvent interactions.30 Thus, there is no evidence for conformational differences in the absence/presence of sucrose.
DISCUSSION
The selection of the right antibody format for a drug candidate is not only a question of targeting the right effector functions suitable for the biological system. Stability of the IgG subclass is also an important factor that can have significant impact both for manufacturing and for drug product formulation. In this study, we have performed extensive comparative analysis of three subclasses of monoclonal IgGs by characterizing their solution behavior, structural, conformational, and colloidal stability. The design of recombinant humanized IgG1, IgG2, and IgG4 with identical CDRs gives unique insights into antibodies of merely different IgG constant composition. The three IgG subclasses were produced using identical purification protocols, and our size-exclusion chromatographic control and glycan analysis reveal samples of high purity and quality. This experimental design hence enables a hitherto high level of qualitative and quantitative comparisons of solution properties of this therapeutically important class of molecules.
Of particular relevance is the improvement of existing and development of novel means to screen various stability parameters in a high-throughput fashion. Several chemical and physical degradation pathways influence protein stability31 and ideally therapeutical antibodies must be investigated by several techniques and under many different formulation conditions. In this study, we reveal how SAXS-based scanning of the solution behavior of IgG subclasses clearly complements the information, which may be retrieved by other well-established biophysical screening methods.
Comparative Analysis of IgG Subclasses Employing Conventional Biophysical Analysis
In this study, we compare how different methods probe different parameters, which are important for the assessment of stability of biopharmaceuticals. The comparison is based on the observations reported here, hence variations may be seen if applying the methods to a different type of proteins or other macromolecules. Some general conclusions can be drawn accordingly. DLS is a very sensitive method to detect the formation of aggregates in a qualitative manner directly in a given formulation as well as study the colloidal stability.32 The thermal stability studied with DSF monitors the conformational stability and interprets the folding–unfolding pathway. It provides an important initial assessment of stability but cannot reliably predict oligomerization, aggregation, and other degradation under long-term storage conditions. Fragmentation is readily monitored by HPLC and LabChip (see Supplementary Material). HPLC is also highly useful for determining the degree of oligomerization and to some extent monitors soluble aggregation. iCE could not provide detailed information about any specific degradation without further characterization, but an overall profile of chemical stability could be sensitively investigated by comparing charge-related heterogeneity (see Supplementary Material).
Subtle differences between subclasses are, however, only revealed when applying all the methods in concert. As an example, acid-induced aggregation was monitored by combining DSF, DLS, and SE-HPLC. Low pH treatment is commonly used in the virus inactivation process or affinity purification of monoclonal antibodies, and we show that IgG subclasses exhibit different aggregation propensity because of differential stability of the corresponding CH2 domain in the Fc region.33 In our studies, DSF data further demonstrated higher conformational stability of IgG1, compared with IgG2 and IgG4, below pH 5.0 and accordingly IgG1 is more resistant to low pH aggregation as observed by DLS (Fig.2c). It is known, in addition, that pH shifts can be used as a chemical stressor, which reproducibly generate artificial aggregates that behave similarly to native ones.34 Antibodies are also prone to fragmentation at acidic pH,35 whereas in our studies all the samples entirely precipitated at pH 3.3 under accelerated storage conditions.
When comparing the large body of data produced in this study, it provides an overall assessment of the conformational and colloidal stability of the individual IgG subclasses. In brief, IgG1 is much more susceptible to fragmentation (Fig.1), whereas possessing a higher overall conformational stability (Fig.2b) and a lower tendency to oligomerize and aggregate (Figs. 1 and 2c). Both conformational and colloidal stability are compromised at the lowest pH value investigated (pH 3.3), particularly for IgG2 and IgG4, whereas the greatest overall stability is achieved around pH 6.5 (Fig.1a; Supplementary Fig. S4e). Sucrose increases the conformational stability and hinders aggregation (Fig.2), whereas fails to rescue the observed chemical degradation (Supplementary Figs. S3b and S4d). Tween is an often used excipient36,37 in the pharmaceutical industry in order to protect against mechanical stress; however, the few samples investigated in the current study do not reveal increased conformational or colloidal stability in the presence of this surfactant at static conditions (not all data are shown).
Complementary Information About IgG Solution Behavior from High-Throughput Compatible SAXS Screening
Small-angle X-ray scattering analysis is not yet a standard method for formulation development in the biopharmaceutical industry, although it is rapidly increasing in use in biomedical research, not the least because of significant advances of dedicated beamlines,38–40 advanced and user-friendly semiautomated software,16,20 and robust in-house equipment.41 SAXS can be employed for detailed advanced structural characterization of the investigated biomacromolecules,42 and can thus also be used for refined structural modeling (not presented in this paper). However, SAXS may also be employed in a high-throughput screening setup, focused on initial primary analysis of basic biophysical characteristics, to which SAXS is very sensitive. The sample consumption for SAXS analysis is continuously diminishing. The current standard screening setup at advanced synchrotron beamlines demands sample volumes of approximately 10 μL,38 and further development toward submicroliter consumption is on-going.43 Also, in-house BioSAXS equipment developed for the analysis of biological samples is becoming increasingly affordable and can be installed with a high level of automation, which attracts more and more interest, also from pharmaceutical companies.
In the current study, we employ SAXS to screen the solution behavior without heat stressing and prior to long-term storage. Interestingly, IgG1 exhibits slightly larger particle size than IgG2 and IgG4 (Fig.4a) in a broad pH range (5.0–8.5), which is in accordance with DLS results (Fig.2c). In addition, an identical solution conformation was identified in this pH range for individual IgG subclass by SAXS, whereas subtle differences among three IgG subclasses were sensitively detected (Fig.3b). Finer differences in the conformational selectivity of IgG subclasses may potentially be modeled in the future. At pH 3.3, dimerization resulting from buffer exchange was sensitively detected for IgG2 and IgG4 before subjecting to heat stress. The samples were freshly prepared and quickly measured by SAXS, thus indicate that the antibody dimerization initiates rapidly after buffer exchange. This may be triggered by acid-induced CH2 unfolding associated with the protonation of specific acidic residues.44 Salt may be an accelerating factor of antibody aggregation at acidic pH by reducing the charge repulsion.45 Sucrose significantly increases physical stability of all three IgG subclasses, likely because of preferential hydration.25 Detailed analysis of the concentration dependence (Figs. 4c and 4d) demonstrated that IgG1 has increased repulsive protein–protein interactions at pH 3.3 with NaCl, which might protect IgG1 against initial dimerization and later unspecific aggregation. This effect is comparable to the general effect of sucrose on all IgG subclasses (Fig.4d). These differences at acidic pH are in agreement with the differences observed by conventional DLS. However, conventional biophysical techniques failed to describe the nature of the oligomeric state and the intermolecular interactions. SAXS is much more sensitive with respect to detection of oligomerization and is therefore used to describe the solution behavior as well as changes at different formulation conditions. In addition, SAXS can provide unique conformational information that potentially enables us to rationalize the functional and physicochemical properties of IgG antibodies.
A Glimpse of the Future Potential of SAXS-Based Formulation Screening
In our study, SAXS reveals differences in solution behavior at initial time points, sensitive to excipient differences and in a subclass specific manner. SAXS might therefore develop into a fast screening method for selection of the most robust molecule because of the detection and description of differences at early time points in contrast to more conventional methods that detected differences only after a given period of time. SAXS may be applied under virtually any relevant formulation conditions without introducing any solvent disturbance or further sample treatment during measurement, which is a powerful asset of the technique. As an example, it is noteworthy that although the presence of Tween complicates several of the standard biophysical analysis types, SAXS readily addresses the solution behavior in the presence of Tween, both in the current and previous studies.10
Although we did not observe significant conformational differences in the pH range of 5.0–8.5 and between NaCl and sucrose, it is possible to investigate the effects of pH and excipients on antibody solution behavior under an appropriate stressing condition, for example, temperature, pH, denaturant, and so on, as exemplified by our studies at acidic pH. Such studies would give further information about the level of robustness of the respective molecules. At the moment, we have not applied SAXS measurements under other stressing conditions, although clearly it is possible to use SAXS for accelerated stability studies. Antibodies are often formulated at very high protein concentrations. It has previously been presented that SAXS readily addresses antibody solution behavior in high-concentration samples.10
CONCLUSIONS
Small-angle X-ray scattering characterization could strongly support selection or development of the therapeutic molecule as well as being part of formulation development of antibodies, or any other therapeutic protein drug candidate. It is emphasized that only primary analysis of the data is necessary to reach the conclusions presented here. For well-behaving samples, the analysis of different formulations can be performed in a semiautomated manner20 using automated sample changer. It is thus possible to use such SAXS-based formulation screening in a high-throughput setup at early stages of formulation development.
Potential transient and/or stable protein–protein interactions at different formulation conditions may also be studied, and it is possible to clearly distinguish levels of oligomerization. An interesting hardware development within solution SAXS analysis is the coupling of FPLC and SAXS measurements,46 which enables quantitative SAXS analysis directly from purified samples, hence addressing, for example, the nature of oligomerization equilibria and offers assessment of the type of, for example, higher-order oligomerization.
If further structural information is requested for particular solution conditions, more advanced data analysis can be applied.47 As an example, a recent study on IgG1 reveals a conformational equilibrium in solution, and a shift toward an “open” conformation was identified under altered solution conditions.48 Likewise, our studies revealed subtle conformational differences among the three IgG subclasses (Fig.3b), which might correlate to their differential physical stability. Further studies by advanced structure modeling are on-going.
In this study, however, it suffices to demonstrate the high degree of complementarity in the information derived from SAXS-based scanning in a comparative study of IgG subclass solution behavior and stability.
Acknowledgments
We thank Zhiru Yang for assistance with protein production and fruitful discussions throughout. We thank Mikkel Melchior Rasmussen for support in biophysical/chemical analysis and Henrik Rahbek Nielsen for glycoanalysis. We appreciate the great support from beamline staff during SAXS beam time at X33 EMBL/DORIS, Hamburg. Funding from the Drug Research Academy, Novo Nordisk A/S, Carlsberg Foundation, and DANSCATT is acknowledged.
Glossary
- CDR
complementarity-determining region
- DLS
dynamic light scattering
- DSF
differential scanning fluorescence
- Dmax
maximal dimension
- Fc
fragment crystallizable
- HMWS
high-molecular-weight species
- I(0)
scattering intensity at zero angle
- iCE
imaged capillary isoelectric focusing
- IgG
immunoglobulin G
- LabChip
automated microfluidic SDS electrophoresis
- LMWS
low-molecular-weight species
- mAb
monoclonal antibody
- P(r)
pair distance distribution function
- Rg
radius of gyration
- Rh
hydrodynamic radius
- SAXS
small-angle X-ray scattering
- SE-HPLC
size-exclusion HPLC
- Tm
thermal transition midpoint
- TNP
2,4,6-trinitrophenol.
This article contains supplementary material available from the authors upon request or via the Internet at http://onlinelibrary.wiley.com/.
REFERENCES
- Jefferis R. Antibody therapeutics: Isotype and glycoform selection. Expert Opin Biol Ther. 2007;7(9):1401–1413. doi: 10.1517/14712598.7.9.1401. [DOI] [PubMed] [Google Scholar]
- Stoop JW, Zegers BJ, Sander PC, Ballieux RE. Serum immunoglobulin levels in healthy children and adults. Clin Exp Immunol. 1969;4(1):101–112. [PMC free article] [PubMed] [Google Scholar]
- Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol. 2010;10(5):301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- Jiskoot W, Beuvery EC, de Koning AA, Herron JN, Crommelin DJ. Analytical approaches to the study of monoclonal antibody stability. Pharm Res. 1990;7(12):1234–1241. doi: 10.1023/a:1015925519154. [DOI] [PubMed] [Google Scholar]
- Rosenberg AS. Effects of protein aggregates: An immunologic perspective. AAPS J. 2006;8(3):E501–507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P, Koza S, Bouvier ES. Size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. J Liquid Chromatogr Relat Technol. 2012;35(20):2923–2950. doi: 10.1080/10826076.2012.743724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J Struct Biol. 2010;172(1):128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod–Debye law. Biopolymers. 2011;95(8):559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Skoda MW, Jacobs RM, Martin RA, Martin CM, Schreiber F. Protein interactions studied by SAXS: Effect of ionic strength and protein concentration for BSA in aqueous solutions. J Phys Chem B. 2007;111(1):251–259. doi: 10.1021/jp0649955. [DOI] [PubMed] [Google Scholar]
- Mosbaek CR, Konarev PV, Svergun DI, Rischel C, Vestergaard B. High concentration formulation studies of an IgG2 antibody using small angle X-ray scattering. Pharm Res. 2012;29(8):2225–2235. doi: 10.1007/s11095-012-0751-3. [DOI] [PubMed] [Google Scholar]
- Jacques DA, Trewhella J. Small-angle scattering for structural biology—Expanding the frontier while avoiding the pitfalls. Protein Sci. 2010;19(4):642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo BK. Antibody humanization by CDR grafting. Methods Mol Biol. 2004;248:135–159. doi: 10.1385/1-59259-666-5:135. [DOI] [PubMed] [Google Scholar]
- Kettleborough CA, Saldanha J, Heath VJ, Morrison CJ, Bendig MM. Humanization of a mouse monoclonal antibody by CDR-grafting: The importance of framework residues on loop conformation. Protein Eng. 1991;4(7):773–783. doi: 10.1093/protein/4.7.773. [DOI] [PubMed] [Google Scholar]
- Kabat EA, Wu T Te, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. Bethesda: Diane Publishing; 1992. [Google Scholar]
- Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun. 2007;355(3):751–757. doi: 10.1016/j.bbrc.2007.02.042. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45(2):342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25(4):495–503. [Google Scholar]
- Round AR, Franke D, Moritz S, Huchler R, Fritsche M, Malthan D, Klaering R, Svergun DI, Roessle M. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. J Appl Crystallogr. 2008;41(5):913–917. doi: 10.1107/S0021889808021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1—Towards automated and web-supported small-angle scattering data analysis. J Appl Crystallogr. 2007;40(s1):s223–s228. [Google Scholar]
- Franke D, Kikhney AG, Svergun DI. Automated acquisition and analysis of small angle X-ray scattering data. Nucl Instrum Meth A. 2012;689(0):52–59. [Google Scholar]
- Salinas BA, Sathish HA, Shah AU, Carpenter JF, Randolph TW. Buffer-dependent fragmentation of a humanized full-length monoclonal antibody. J Pharm Sci. 2010;99(7):2962–2974. doi: 10.1002/jps.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JY, Janis LJ. Influence of pH, buffer species, and storage temperature on physicochemical stability of a humanized monoclonal antibody LA298. Int J Pharm. 2006;308(1–2):46–51. doi: 10.1016/j.ijpharm.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Kaushik JK, Bhat R. Why is trehalose an exceptional protein stabilizer? An analysis of the thermal stability of proteins in the presence of the compatible osmolyte trehalose. J Biol Chem. 2003;278(29):26458–26465. doi: 10.1074/jbc.M300815200. [DOI] [PubMed] [Google Scholar]
- Xie G, Timasheff SN. Mechanism of the stabilization of ribonuclease A by sorbitol: Preferential hydration is greater for the denatured then for the native protein. Protein Sci. 1997;6(1):211–221. doi: 10.1002/pro.5560060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timasheff SN. Protein-solvent preferential interactions, protein hydration, and the modulation of biochemical reactions by solvent components. Proc Natl Acad Sci USA. 2002;99(15):9721–9726. doi: 10.1073/pnas.122225399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Bautista R, Yu K, Zapata GA, Mulkerrin MG, Chamow SM. Influence of histidine on the stability and physical properties of a fully human antibody in aqueous and solid forms. Pharm Res. 2003;20(12):1952–1960. doi: 10.1023/b:pham.0000008042.15988.c0. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Ito T, Endo R, Nakagawa K, Sawa E, Wakamatsu K. Influence of pH on heat-induced aggregation and degradation of therapeutic monoclonal antibodies. Biol Pharm Bull. 2010;33(8):1413–1417. doi: 10.1248/bpb.33.1413. [DOI] [PubMed] [Google Scholar]
- Mylonas E, Svergun DI. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J Appl Crystallogr. 2007;40(s1):s245–s249. [Google Scholar]
- Timasheff SN. Solvent stabilization of protein structure. Methods Mol Biol. 1995;40:253–269. doi: 10.1385/0-89603-301-5:253. [DOI] [PubMed] [Google Scholar]
- Wu D, Minton AP. Quantitative characterization of the interaction between sucrose and native proteins via static light scattering. J Phys Chem B. 2013;117(1):111–117. doi: 10.1021/jp308880v. [DOI] [PubMed] [Google Scholar]
- Wang W, Singh S, Zeng DL, King K, Nema S. Antibody structure, instability, and formulation. J Pharm Sci. 2007;96(1):1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- Lehermayr C, Mahler HC, Mader K, Fischer S. Assessment of net charge and protein–protein interactions of different monoclonal antibodies. J Pharm Sci. 2011;100(7):2551–2562. doi: 10.1002/jps.22506. [DOI] [PubMed] [Google Scholar]
- Hari SB, Lau H, Razinkov VI, Chen S, Latypov RF. Acid-induced aggregation of human monoclonal IgG1 and IgG2: Molecular mechanism and the effect of solution composition. Biochemistry. 2010;49(43):9328–9338. doi: 10.1021/bi100841u. [DOI] [PubMed] [Google Scholar]
- Potty ASP, Xenopoulos A. Stress-induced antibody aggregates. BioProcess Int. 2013;11(3):44–52. [Google Scholar]
- Vlasak J, Ionescu R. Fragmentation of monoclonal antibodies. mAbs. 2011;3(3):253–263. doi: 10.4161/mabs.3.3.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang YJ, Wang DQ. Dual effects of Tween 80 on protein stability. Int J Pharm. 2008;347(1–2):31–38. doi: 10.1016/j.ijpharm.2007.06.042. [DOI] [PubMed] [Google Scholar]
- Chou DK, Krishnamurthy R, Randolph TW, Carpenter JF, Manning MC. Effects of Tween 20 and Tween 80 on the stability of Albutropin during agitation. J Pharm Sci. 2005;94(6):1368–1381. doi: 10.1002/jps.20365. [DOI] [PubMed] [Google Scholar]
- Blanchet CE, Zozulya AV, Kikhney AG, Franke D, Konarev PV, Shang W, Klaering R, Robrahn B, Hermes C, Cipriani F, Svergun DI, Roessle M. Instrumental setup for high-throughput small- and wide-angle solution scattering at the X33 beamline of EMBL Hamburg. J Appl Crystallogr. 2012;45(3):489–495. [Google Scholar]
- Pernot P, Round A, Barrett R, Maria Antolinos A De, Gobbo A, Gordon E, Huet J, Kieffer J, Lentini M, Mattenet M, Morawe C, C Mueller-Dieckmann, Ohlsson S, Schmid W, Surr J, Theveneau P, Zerrad L, McSweeney S. Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J Synchrotron Radiat. 2013;20(Pt 4):660–664. doi: 10.1107/S0909049513010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C, Svergun D. Upgrade of the small-angle X-ray scattering beamline X33 at the European molecular biology laboratory, Hamburg. J Appl Crystallogr. 2007;40(s1):s190–s194. [Google Scholar]
- Pedersen J. A flux- and background-optimized version of the NanoSTAR small-angle X-ray scattering camera for solution scattering. J Appl Crystallogr. 2004;37(3):369–380. [Google Scholar]
- Graewert MA, Svergun DI. Impact and progress in small and wide angle X-ray scattering (SAXS and WAXS. Curr Opin Struct Biol. 2013;23(5):748–754. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Møller M, Nielsen SS, Ramachandran S, Li Y, Tria G, Streicher W, Petoukhov MV, Cerione RA, Gillilan RE, Vestergaard B. Small angle X-ray scattering studies of mitochondrial glutaminase C reveal extended flexible regions, and link oligomeric state with enzyme activity. PloS one. 2013;8(9):e74783. doi: 10.1371/journal.pone.0074783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latypov RF, Hogan S, Lau H, Gadgil H, Liu D. Elucidation of acid-induced unfolding and aggregation of human immunoglobulin IgG1 and IgG2 Fc. J Biol Chem. 2012;287(2):1381–1396. doi: 10.1074/jbc.M111.297697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrih NP, Anderluh G, Macek P, Chalikian TV. Salt-induced oligomerization of partially folded intermediates of equinatoxin II. Biochemistry. 2004;43(29):9536–9545. doi: 10.1021/bi049616h. [DOI] [PubMed] [Google Scholar]
- David G, Perez J. Combined sampler robot and high-performance liquid chromatography: A fully automated system for biological small-angle X-ray scattering experiments at the synchrotron SOLEIL SWING beamline. J Appl Crystallogr. 2009;42(5):892–900. [Google Scholar]
- Blanchet CE, Svergun DI. Small-angle X-ray scattering on biological macromolecules and nanocomposites in solution. Annu Rev Phys Chem. 2013;64:37–54. doi: 10.1146/annurev-physchem-040412-110132. [DOI] [PubMed] [Google Scholar]
- Lilyestrom WG, Shire SJ, Scherer TM. Influence of the cosolute environment on IgG solution structure analyzed by small-angle X-ray scattering. J Phys Chem B. 2012;116(32):9611–9618. doi: 10.1021/jp303839t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.