Figure 6.
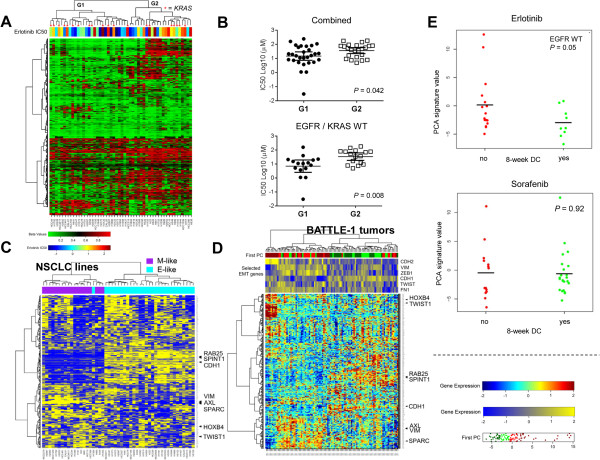
Extent of methylated SRAMs associated with erlotinib response in cell lines and patients in BATTLE-1 trial. A) Hierarchical clustering of EMT-SRAMs in 52 wild-type (WT) EGFR NSCLC cell lines, with erlotinib IC50 levels overlying individual cell lines. G1 and G2 represent the two cell lines clustered by SRAM methylation. KRAS mutant cells are represented by asterisks. B) Erlotinib IC50 values of G1 vs G2 clusters for the combined cell lines, and the WT EGFR/KRAS cells. G2 cluster had significantly higher IC50 values than G1 cluster. Error bars indicate average and 95% confidence interval. C) Hierarchical clustering of expression of SRAMs in the panel of cell lines with RPPA data for E-cadherin (49) clusters of E and M cells. Specific gene markers that relate to EMT are on the right. D) Hierarchical clustering based on first principle component analysis (PCA) of SRAM expression signature in the tumor biopsies from patients in BATTLE-1 trial clusters tumors into groups that resemble the separation seen in the cell lines. E) Eight-week disease control (DC) of patients treated with erlotinib or sorafenib based on the first PCA separation. EGFR mutant tumors were excluded.
