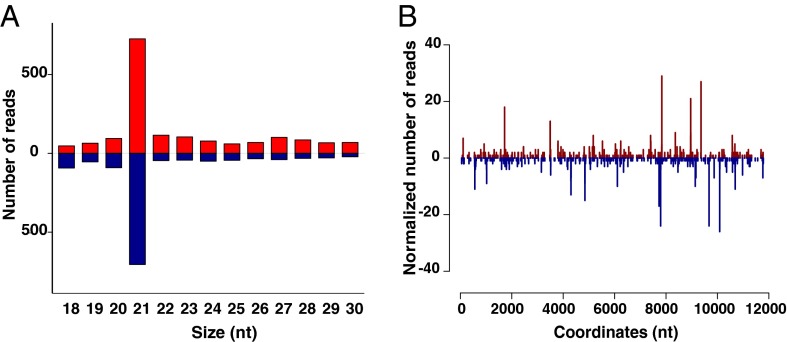
Fig. 3.
Exogenous siRNA pathway is functionally active but not antiviral in the An. gambiae midgut. (A) Size distribution of viRNAs in An. gambiae mosquitoes 3 d after an ONNV-infective blood meal, when viral replication is restricted to the midgut. Positive numbers on the y axis indicate the frequency of viRNAs mapping to the ONNV genome (red bars), and negative numbers on the y axis indicate the frequency of viRNAs mapping to the ONNV antigenome (blue bars). (B) Frequency distribution of loci in the ONNV genome generating 21-nt viRNA (5′ to 3′; viRNAs starting with the 5′ terminal nucleotide) or antigenome (3′ to 5′; viRNAs starting with the 3′ terminal nucleotide), starting at nucleotide 1 on the x axis. Numbers on the y axis correspond to the frequencies of viRNAs mapping to the ONNV genome (positive) or antigenome (negative). Blue peaks indicate loci of viRNAs mapping to the ONNV genome, and red peaks indicate loci of viRNAs mapping to the ONNV antigenome. Results shown are representative of two replicates.