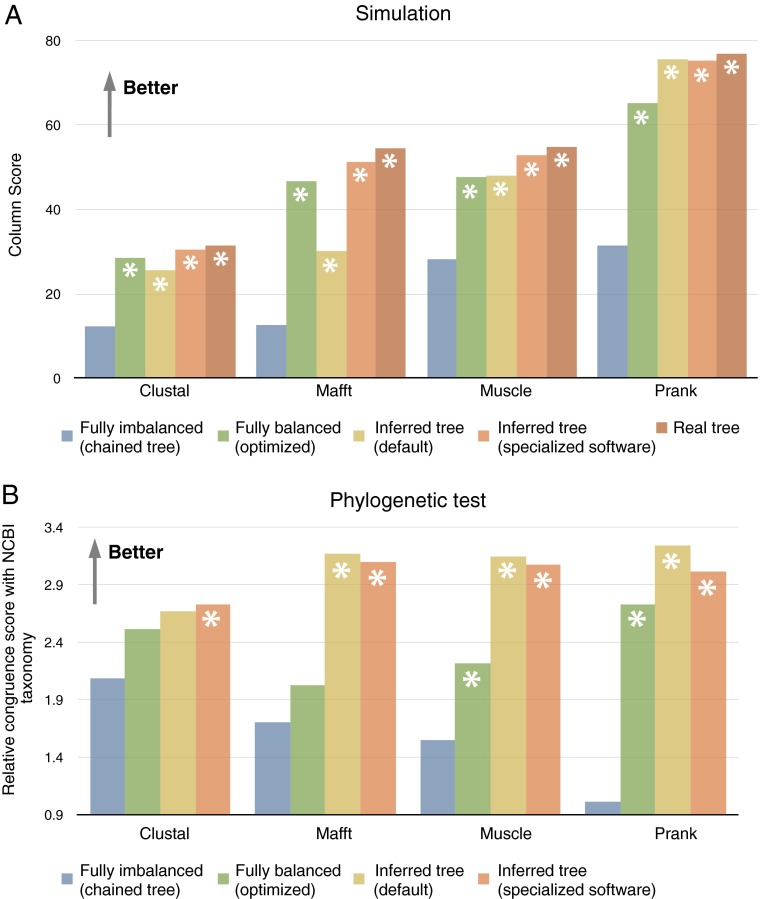
Fig. 1.
Evaluation of alignments reconstructed with various aligners and guide tree methods. (A) Average true column score over 113 simulated datasets of 1,024 sequences. (B) Average consistency with the National Center for Biotechnology Information taxonomy over 106 sets of 1,024 biological sequences. Note that the real tree is unknown for empirical data. With fully imbalanced trees as input guide tree, Prank failed to reconstruct alignments in 38 empirical data problem instances; results reported in B are thus based on the remaining 68 alignments. Significant difference from fully imbalanced guide trees is indicated with an asterisk (Wilcoxon double-sided test, P < 0.001). All data available at dx.doi.org/10.5061/dryad.4r5b8.