Significance
In this study, we have identified an important role of mammalian Lnp1 (mLnp1) in stabilizing nascent three-way ER junctions. When new junctions acquire mLnp1, they tend to remain stable within the ER network, whereas the nascent junctions that fail to acquire mLnp1 preferentially undergo ring closure. Our findings provide insights into the function of mLnp1.
Keywords: endoplasmic reticulum, organelle morphogenesis, three-way junction, Lunapark, membrane remodeling
Abstract
The endoplasmic reticulum (ER) consists of a polygonal network of sheets and tubules interconnected by three-way junctions. This network undergoes continual remodeling through competing processes: the branching and fusion of tubules forms new three-way junctions and new polygons, and junction sliding and ring closure leads to polygon loss. However, little is known about the machinery required to generate and maintain junctions. We previously reported that yeast Lnp1 localizes to ER junctions, and that loss of Lnp1 leads to a collapsed, densely reticulated ER network. In mammalian cells, only approximately half the junctions contain Lnp1. Here we use live cell imaging to show that mammalian Lnp1 (mLnp1) affects ER junction mobility and hence network dynamics. Three-way junctions with mLnp1 are less mobile than junctions without mLnp1. Newly formed junctions that acquire mLnp1 remain stable within the ER network, whereas nascent junctions that fail to acquire mLnp1 undergo rapid ring closure. These findings imply that mLnp1 plays a key role in stabilizing nascent three-way ER junctions.
The endoplasmic reticulum (ER) is a large, membrane-bounded organelle responsible for a number of essential cellular processes in eukaryotic cells (1). Typically, the ER forms a continuous polygonal membrane network composed of interconnected sheets and tubules that spreads throughout the cytoplasm. Several key evolutionarily conserved factors involved in the formation and maintenance of the ER structure have been identified. Reticulons and DP1(deleted in polyposis)/Yop1(YIP one partner) are integral membrane proteins predominantly associated with the peripheral ER (2, 3). They are thought to contribute local positive curvature to the ER membrane and thereby help to generate and stabilize ER tubules as well as the tight bends at the edges of ER sheets (4, 5). The dynamin-like GTPase atlastin is enriched at the three-way junctions of the ER tubular network. Atlastin drives homotypic ER membrane fusion (6, 7), an activity that is needed to establish ER tubule interconnections and is therefore essential for the formation of a typical ER network. The medical importance of these ER shaping proteins is highlighted by the neurologic disorder hereditary spastic paraplegia (HSP). Mutations in atlastin, reticulon, DP1/REEP1, or their family members lead to alterations in ER morphology that are believed to be the primary pathological defects underlying HSP (8).
We recently identified Lnp1 (Lunapark), a member of the conserved Lunapark protein family that is required for the maintenance of the ER network in yeast cells (9). When LNP1 function is disrupted, yeast cells exhibit a collapsed cortical ER network accompanied by large sectors of the cortex devoid of ER (9). The Lnp1 protein resides at three-way junctions of the ER network in yeast and mammalian cells. The Lunapark protein family is defined by two transmembrane domains and a zinc finger motif that is essential for ER morphogenesis in yeast. Genetic analysis indicates that Lnp1 acts in synergy with the reticulons and Yop1 but in antagonism to Sey1 (synthetic enhancement of YOP1), the atlastin homolog in yeast.
The fine structure of the peripheral ER network of mammalian cells is architecturally similar to the cortical ER network in yeast, consisting of a polygonal network of sheets and tubules. This interconnected ER network undergoes constant remodeling through tubule extension, tubule retraction, tubule–tubule fusion, tubule sliding, and ring closure (10). These events are generally accompanied by junction dynamics. New junctions are generated when a tubule fuses to the side of another tubule. Preexisting junctions can change their position by sliding along one of the intersecting tubules. Junction sliding can lead to loss of one leg of a polygon or complete loss of an entire polygon and its associated junctions, a process known as ring closure. At steady state, the loss of junctions by ring closure balances new junction formation (10). Thus, junctions act as unique mobile elements within the ER network and junction dynamics enable reorganization of the ER network morphology over time. Nonetheless, the machinery involved in junction dynamics remains unclear. Our previous observation that Lnp1 resides at the three-way junctions of the ER network (9) suggests that Lnp1 might function in ER morphogenesis specifically through junction-mediated activity. The lnp1∆ mutant of yeast loses its typical ER distribution pattern at the cortex. Loss of Lnp1 might result in aberrant junction dynamics, leading to the collapsed ER morphology and cortical regions devoid of ER observed in lnp1∆ mutant cells (9).
In this study, we demonstrate heterogeneity among three-way junctions in mammalian cells with regards to Lnp1. Junctions with mammalian Lnp1 (mLnp1) are less mobile than junctions lacking mLnp1. Moreover, the presence of mLnp1 appears to be correlated with stabilization of nascent junctions. When new junctions acquire mLnp1, they tend to remain stable within the ER network, whereas the nascent junctions that fail to acquire mLnp1 preferentially undergo ring closure. We propose that mLnp1 functions in the maintenance of ER morphology through stabilization of junctions.
Results
mLnp1 Is Required for the Maintenance of ER Structure in Mammalian Cells.
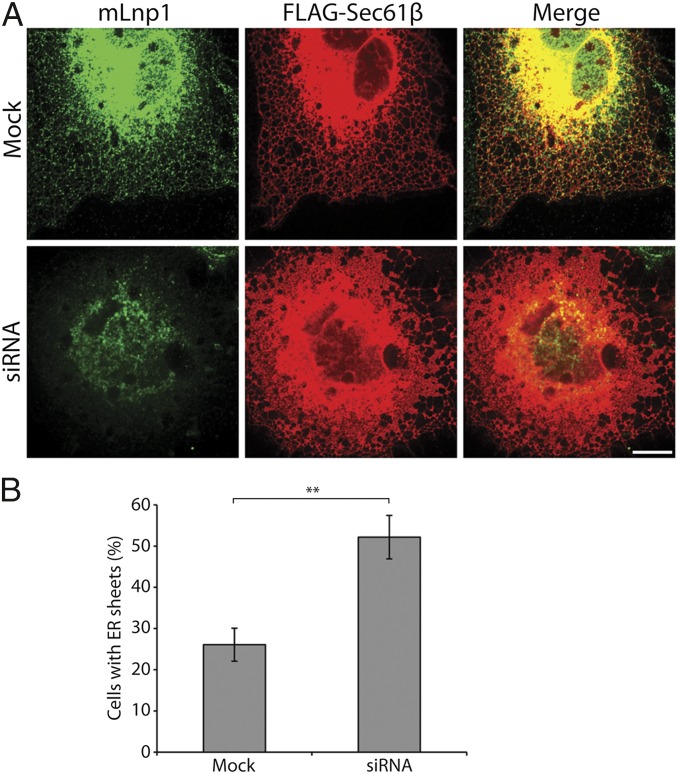
We previously showed that Lnp1 plays an important role in maintaining the ER network in yeast (9). Specifically, we found that the disruption of LNP1 leads to a collapsed, more densely reticulated cortical ER network (9). As Lunapark homologs localize to the three-way junctions of the ER network in yeast and mammalian cells and share conserved structural features (9), we anticipated that mLnp1 would also play a role in ER maintenance. To test this prediction, we depleted mLnp1 by transfecting COS-7 cells with an siRNA duplex or two different shRNAs (Lnp1A and Lnp1B) that target mLnp1 mRNA. Western blot analysis revealed that mLnp1 was reduced 96% with siRNA (Fig. S1), 79% with shRNA Lnp1A, and 77% with shRNA Lnp1B (Fig. S2A). In all three cases, depletion of mLnp1 led to an increase of ER sheets with a reduction in tubular structures (Fig. 1 and Fig. S2 B and C), suggesting a shift of the ER morphology from a tubular to more sheet-like organization. Quantification of the effects of mLnp1 depletion on ER morphology showed that 51.2% of the siRNA-treated cells displayed an increase in ER sheets, compared with 26.1% in mock-treated cells (Fig. 1B). Similar results were observed when Lnp1A and Lnp1B were used to deplete mLnp1 (Fig. S2C). Thus, mLnp1 appears to play a role in shaping or maintaining the morphology of the ER network in mammalian cells.
Fig. 1.
Depletion of mLnp1 shifts the morphology of the ER from tubular to more sheet-like. (A) COS-7 cells were cotransfected with FLAG-Sec61β and siRNA directed against mLnp1 or mock-treated. Cells were immunostained for mLnp1 and FLAG-Sec61β. (Scale bar: 10 μm.) (B) The percentage of cells with a sheet-like ER morphology was quantified. Cells with ER sheets were defined as those in which the area of the sheets was more than 50% of the total area of the ER network. Error bars are SEM for three separate experiments. n = 297 cells for mock transfection, n = 349 cells for LNP1 siRNA transfected cells (**P < 0.01, Student t test).
mLnp1 Is Not Present at All Junctions of the ER Network.
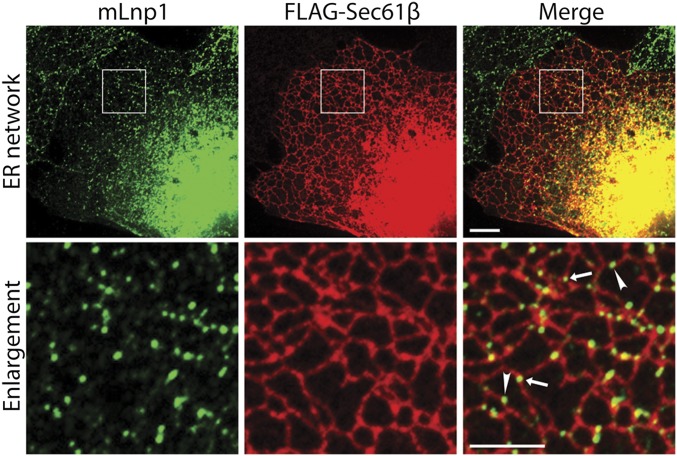
By using COS-7 cells transfected with mLnp1-GFP (green fluorescent protein) and markers of the ER network, we previously reported that mLnp1-GFP localizes to the three-way junctions of the ER (9). When we performed indirect immunofluorescence in untransfected COS-7 cells by using an antibody directed against mLnp1, endogenous mLnp1 showed the same localization pattern as transfected mLnp1-GFP. In fixed COS-7 cells, 72.4% of mLnp1 puncta localized to the three-way junctions of the ER, whereas 27.6% of the mLnp1 puncta were found on tubules (Fig. 2). Additionally, when we quantified the number of junctions with and without mLnp1, we found that mLnp1 was not present at all junctions. mLnp1 was found at 51.7% of the ER junctions in COS-7 cells and absent from 48.3% of the junctions.
Fig. 2.
mLnp1 localizes to a subset of the three-way junctions in the ER network and to puncta on ER tubules. COS-7 cells were transfected with FLAG-Sec61β that was immunostained to visualize the ER network, whereas mLnp1 was immunostained with anti-Lunapark antibody. The boxed area (Top) is enlarged below. Arrow indicates mLnp1 punctum on junctions, arrowhead indicates mLnp1 punctum on a tubule. (Scale bars: Top, 10 μm; Bottom, 5 μm.)
Atlastin is also associated with three-way junctions (11), and, in yeast, the atlastin homolog Sey1 partially colocalizes with Lnp1 at three-way junctions (9). We found that human Atl3 (atlastin-3) tagged with GFP (GFP-Atl3) partially colocalized with mLnp1 in COS-7 cells (Fig. S3). Approximately 80.5% of the mLnp1 puncta colocalized with GFP-Atl3 puncta, whereas 82.1% of the GFP-Atl3 puncta colocalized with mLnp1 puncta (Fig. S3). As atlastin is a dynamin-like GTPase that facilitates homotypic ER membrane fusion (6, 7), and Lnp1 and Sey1 act antagonistically to balance polygonal ER network formation in yeast (9), we examined whether Lnp1 affects the membrane fusion and GTPase activity of atlastin. By using a liposome-based fusion assay, we found that the soluble domain of human or Drosophila Lnp1 had no significant effect on the fusion driven by Drosophila atlastin (Fig. S4 A–C) or GTPase activities of human atlastin-1 (Fig. S4D). These findings suggest that Lnp1 does not control ER morphology maintenance by directly affecting atlastin-mediated ER membrane fusion, although we cannot exclude the possibility that this potential function of Lnp1 requires the transmembrane domains.
The mLnp1 at ER Junctions Is Not Highly Mobile.
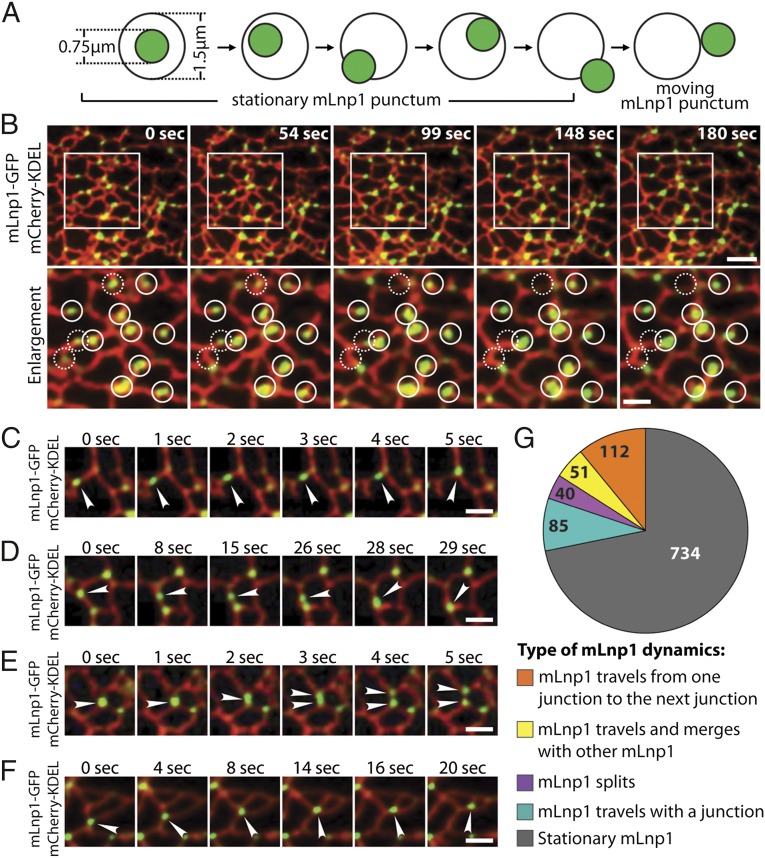
Three-way junctions are dynamic elements within the ER network. New junctions are formed by tubule–tubule fusion, and preexisting junctions may slide along tubules or be lost as an ER polygon undergoes ring closure (10). Because mLnp1 largely localizes to junctions, one might expect that mLnp1 will undergo dynamic changes as the ER reorganizes its network of tubules and junctions. We addressed this question in live COS-7 cells. To assess the dynamics of mLnp1, we first established criteria to determine if mLnp1 puncta are stationary or mobile. In COS-7 cells, the ER network was marked with mCherry-KDEL, and mLnp1 was tagged with GFP. mLnp1-GFP at junctions was visualized as puncta with an average apparent diameter of 0.75 μm (Fig. 3A). Around each mLnp1-GFP punctum, a center-aligned 1.5-μm-diameter circle was established as a restricted region. By using time-lapse movies, stationary mLnp1 puncta were defined as those that do not move beyond the circle during 180 s, whereas mobile puncta were defined as those that move entirely out of the circle in this time period (Fig. 3A). This analysis revealed that mLnp1 puncta generally remained stationary at junctions. Despite the changes in the surrounding ER structure, most mLnp1 puncta did not leave the junctions where they originally resided, and did not move out of the restricted region in 180 s (Fig. 3B, solid circles, and Movie S1). Moreover, the junctions marked by these mLnp1 puncta appeared to be stationary as well. These data suggest a tight association of mLnp1 with ER junctions. Nonetheless, a small portion of mLnp1 puncta exhibited mobile behavior, as they moved out of the restricted region during 180 s (Fig. 3B, dashed circles, and Movie S1).
Fig. 3.
Dynamics of mLnp1. (A) A diagram showing how mLnp1 dynamics were determined. The average diameter of mLnp1 puncta (green) was approximately 0.75 μm, and a 1.5-μm-diameter circle was used to mark the restricted region. Stationary mLnp1 puncta are defined as those puncta that do not move out of the circle during 180 s. Mobile mLnp1 puncta are defined as those puncta that move entirely out of the circle. (B) The dynamics of mLnp1 puncta were monitored by using mLnp1-GFP. The ER tubules were marked by mCherry-KDEL. The boxed area (Top) is enlarged below. Solid circles (diameter, 1.5 μm) mark the original position of stationary mLnp1 puncta. Dashed circles (diameter, 1.5 μm) mark the original position of mobile mLnp1 puncta. (Scale bars: Top, 4 μm; Bottom, 2 μm.) (C–F) The dynamic properties of mLnp1 puncta are shown through time-lapse imaging. (C) An mLnp1 punctum moves from one junction to another junction along an ER tubule. (D) An mLnp1 punctum merges with another mLnp1 puncta. (E) An mLnp1 punctum splits into two puncta. (F) An mLnp1 punctum moves with a junction as it undergoes sliding. (Scale bars: 2 μm.) Arrowheads in C–F point to mobile mLnp1 puncta. (G) Quantification of mLnp1 dynamics (n = 1,022 mLnp1 puncta).
Among the mobile mLnp1 puncta, four categories of motion were observed (Fig. 3 C–F). First, an mLnp1 punctum moves from one junction to the next along an ER tubule (Fig. 3C and Movie S2). Second, if mLnp1 puncta are present at two adjacent junctions, one mLnp1 punctum can merge with its neighbor to form a single punctum (Fig. 3D and Movie S3). In both events, the mobile mLnp1 punctum appeared to move as an intact unit, leaving behind an mLnp1-free junction, without affecting the dynamics of these junctions. Third, the mLnp1 punctum at a junction may split into two independent puncta, with one punctum remaining at the original junction and the other moving to an adjacent junction (Fig. 3E and Movie S4). This event may serve to balance events that lead to the merging mLnp1 puncta. Fourth, the mLnp1 punctum travels with a junction that is undergoing sliding (Fig. 3F and Movie S5). During its travels, the mLnp1 follows the sliding pathway at all times and does not disassociate from the junction.
When we analyzed the dynamics of 1,022 mLnp1-GFP puncta by using the criteria illustrated in Fig. 3A, 734 were found to be stationary, whereas 288 were mobile (Fig. 3G). The preponderance of stationery puncta in the ER network may have an important effect on the dynamics of the junctions. The movement of mLnp1 between junctions, as described in Fig. 3 C–E, could explain why, at any given time, a fraction of mLnp1 puncta are observed on tubules, by live cell imaging and immunofluorescence (see Fig. 2). Although this small population of mLnp1 puncta might be in transit along a tubule (Fig. 2), puncta preferentially move to, or travel with, junctions (Fig. 3 C–F), further supporting a role for mLnp1 at junctions.
Junctions with mLnp1 Are Less Mobile than Junctions Without mLnp1.
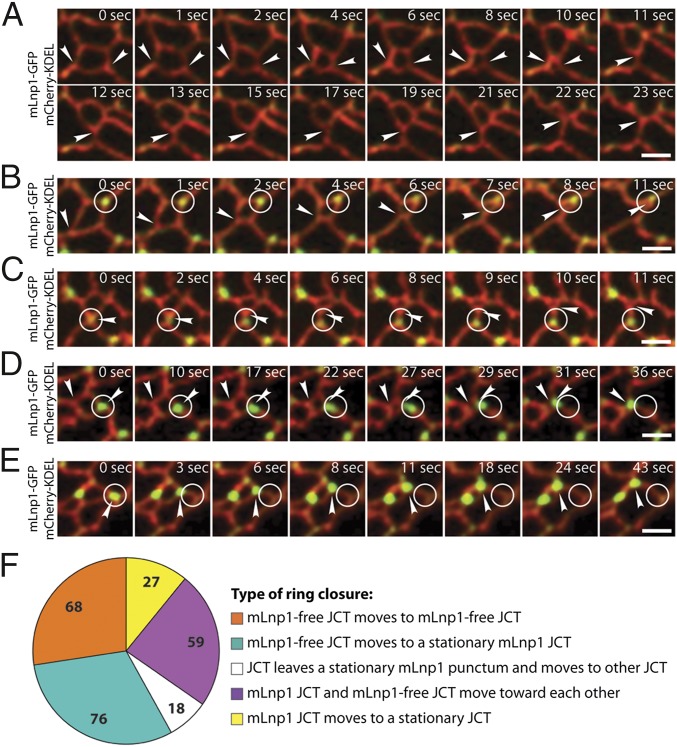
The observation that mLnp1 resides at or travels between junctions suggests that mLnp1 exerts its effect on ER morphology by impacting the three-way junctions of the network. Although the dynamics of ER morphology are achieved in part by junction-mediated events such as junction sliding and ring closure (10), and mLnp1 preferentially associates with junctions (Fig. 3 B and G), approximately half of all junctions lack mLnp1 (Fig. 2). To begin to address the relationship of mLnp1 to junction dynamics, we examined the motion of preexisting junctions that undergo ring closure. During ring closure, junctions slide along one of the intersecting tubules to decrease the size of the polygon until it vanishes (10). In COS-7 cells transfected with mCherry-KDEL and mLnp1-GFP, we identified five categories of ring closure based on the motions of polygon junctions. First, a polygon that undergoes ring closure contains only mLnp1-free junctions. Ring closure is achieved through the sliding of these mLnp1-free junctions (Fig. 4A and Movie S6). Second, a polygon contains junctions with mLnp1 and junctions without mLnp1. Junctions with mLnp1 remain stationary. The mLnp1-free junctions slide along the neighboring tubules to complete the process of ring closure (Fig. 4B and Movie S7). Third, on rare occasions, a polygon contains a four-way junction with an mLnp1 punctum. The four-way junction splits into two three-way junctions. The junction that retains mLnp1 remains stationary, whereas the junction that loses mLnp1 slides away to complete a ring closure (Fig. 4C and Movie S8). Four, all junctions with mLnp1 remain stationary during ring closure. Sometimes ring closures can occur through the sliding of junctions that contain mLnp1 and those that do not (Fig. 4D and Movie S9). Finally, some ring closures are achieved by the sliding of junctions that contain mLnp1 (Fig. 4E and Movie S10). When we examined 248 ring closure events for the movement of polygon junctions, 162 (65.3%) were found to fall into categories one, two, or three, whereas 86 (34.7%) were in categories four and five (Fig. 4F). These results suggest that not all polygon junctions freely slide during the process of ring closure. mLnp1-free junctions tend to be mobile, because the majority of ring closure events occur through the sliding of junctions lacking mLnp1. By contrast, only a minor portion of ring closures could be attributed to the movement of junctions with mLnp1. These data suggest that junctions with mLnp1 may be more stable within the ER network than those lacking mLnp1.
Fig. 4.
Preexisting mLnp1-free junctions undergo more movements leading to ring closure. (A–E) The various types of ring closure events that we observed are shown through time-lapse imaging. The ER tubules are marked by mCherry-KDEL. mLnp1 puncta were monitored by using mLnp1-GFP. (A) An mLnp1-free junction moves to another mLnp1-free junction. (B) An mLnp1-free junction moves to a stationary mLnp1 junction. (C) A junction leaves a stationary mLnp1 punctum and moves to another junction. (D) An mLnp1 junction and an mLnp1-free junction move toward each other. (E) An mLnp1 junction moves to a stationary junction. The arrowhead points to a moving junction. The circle marks the original position of an mLnp1 punctum. (Scale bars: 2 μm.) (F) Quantification of the various types of ring closure events (n = 248 ring closures).
To further characterize the motility of junctions, we measured their velocities. Calculations of the trajectory and the total distance that a junction travels are illustrated in Fig. S5A. The average velocities are calculated by dividing the total distance by the total elapsed time. We obtained velocity measurements for 704 junctions, which were graphed in a histogram (Fig. S5B). We defined 0–200 nm/s as a low velocity and 200–500 nm/s as a high velocity. Our observations revealed that most junctions (512 of 704) move at low velocities, and only a small portion of junctions (192 of 704) move at high velocities (Fig. S5 B and C). However, the velocity distribution profile of junctions with mLnp1 is not the same as that of mLnp1-free junctions. Among 355 junctions with mLnp1, 286 (80.6%) move with velocities of 0–200 nm/s and 69 (19.4%) move with velocities of 200–500 nm/s. In contrast, 226 of 349 (64.8%) mLnp1-free junctions move with velocities of 0–200 nm/s and 123 (35.2%) move at 200–500 nm/s. These data support the hypothesis that junctions with mLnp1 tend to be less mobile than mLnp1-free junctions.
The Presence of mLnp1 Correlates with the Stabilization of Nascent Junctions.
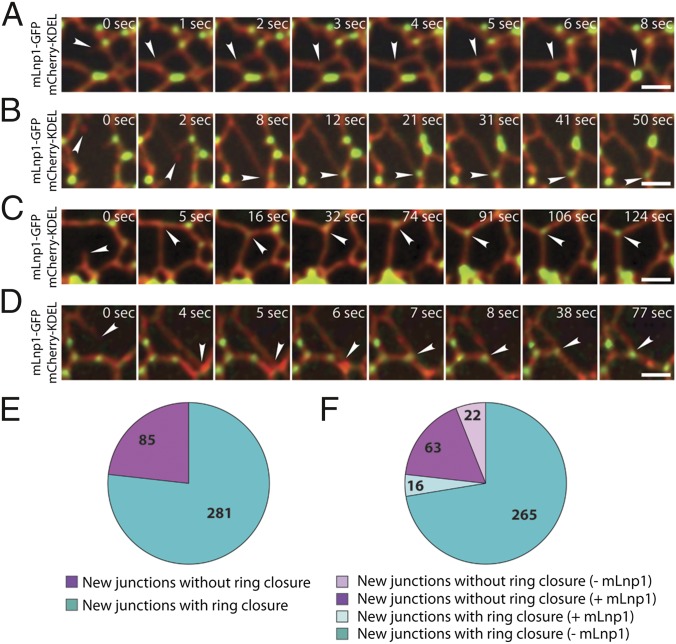
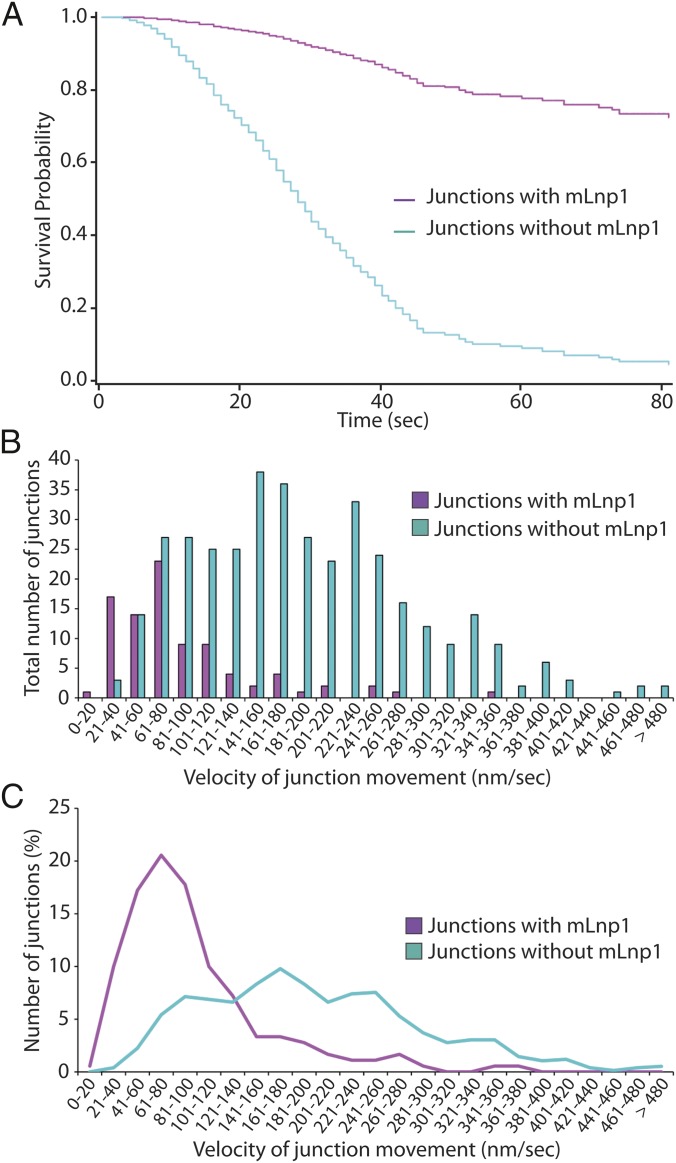
A new junction is generated when the tip of an ER tubule fuses to the side of another tubule. The observation that preexisting junctions with and without mLnp1 exhibit motility differences prompted us to examine how nascent junctions behave after they are generated, and the relationship of mLnp1 to the fate of nascent junctions. We analyzed the dynamics of nascent junctions over a 90–180-s time period and assessed their stability. We defined unstable nascent junctions as those junctions that are lost as a result of ring closure during the period of observation, as indicated in Fig. 5A and Fig. S6A (also see Movie S11). Stable nascent junctions are defined as those that are not lost as a result of ring closure and maintain their structure until the end of the observation period (provided that we were able to observe the junction for at least 30 s after formation; Fig. 5 B–D and Movie S12). By using these criteria, we found that the majority of nascent junctions were not stable. Among the 366 nascent junctions examined, 281 (76.8%) were lost as a result of ring closure at times from 1 to 85 s after formation, with a mean period of 26 s (Fig. S6B), whereas 85 (23.2%) appeared to be stable over the period of observation (Fig. 5E). Moreover, 265 of the 281 unstable junctions did not acquire mLnp1 over their lifetime. Thus, it seems likely that the absence of mLnp1 correlates with instability of nascent junctions through ring closure. This is consistent with our analysis of preexisting mLnp1-free junctions (Fig. 4). Intriguingly, 63 of 85 stable junctions acquired mLnp1 soon after they were generated. Despite occasional sliding or slow movement, these junctions persisted over the time period of observation (Fig. 5 B–D). By contrast, of the 281 junctions that underwent ring closure, only 16 acquired mLnp1 (Fig. 5F). These results indicate that the acquisition of mLnp1 strongly correlates with the stabilization of nascent junctions. This correlation was further confirmed by the analysis of junction survival probability. Nascent junctions that did not acquire mLnp1 displayed an extremely low survival probability. When they have formed, they slide away from their site of origin to the adjacent junction, and ultimately disappear as a result of ring closure (Fig. 5A). Within 85 s after new junctions were generated, only 5% of the junctions that had not acquired mLnp1 survived. However, among junctions that acquired mLnp1, 73% survived (Fig. 6A). Thus, the presence of mLnp1 appears to enhance the survival probability of nascent junctions, suggesting an important role of mLnp1 in the stabilization of nascent junctions.
Fig. 5.
The presence of mLnp1 is correlated with stabilization of newly formed junctions. (A) Time course of a newly formed junction undergoing ring closure. The ER tubules were marked by mCherry-KDEL. The arrowhead points to a new junction lacking mLnp1-GFP. (B–D) Time course of a new junction that does not undergo ring closure during the indicated time period. These junctions remain stable until the end of the observation period of at least 30 s. The ER tubules were marked by mCherry-KDEL. The arrowhead in A points to a new junction that did not acquire mLnp1-GFP, and the arrowheads in B–D point to new junctions that acquired mLnp1-GFP. (Scale bars: 2 μm.) (E) Quantification of stability of new junctions. In a total of 366 new junctions, 85 are stable (magenta) and 281 undergo ring closure (cyan). (F) Of the 85 stable junctions analyzed, 63 acquired mLnp1 (magenta). Of the 281 junctions that underwent ring closure, 265 lacked mLnp1.
Fig. 6.
The presence of mLnp1 enhances the survival probability of nascent junctions, and reduces their velocities. (A) The survival probability was calculated for newly formed junctions that did (magenta) or did not (cyan) acquire mLnp1 during the observation period. The solid lines represent the estimated survival function and the symbols represent data values. Time was measured from when one tubule was seen to fuse with another, generating a new junction. Nascent junctions that had not undergone ring closure at the end of the 180-s observation period were considered censored. (B) Quantification of the velocities of newly formed junctions. n = 90 for junctions with mLnp1 (magenta); n = 378 for junctions lacking mLnp1 (cyan). (C) A trendline of the velocities of newly formed junctions based on the data in B.
We also calculated the velocities of nascent junctions as we did for preexisting junctions. Time 0 was defined as the time when a branching tubule fused with a preexisting tubule to generate a new junction. The “end” time point was when ring closure was completed or when the observation period ended (Fig. S6A). Our analysis revealed that 93.3% (84 of 90) of the nascent junctions marked by mLnp1 moved at 0–200 nm/s, whereas only 6.7% (6 of 90) moved at 200–500 nm/s (Fig. 6 B and C). By contrast, 58.7% (222 of 378) of the nascent junctions lacking mLnp1 had velocities of 0–200 nm/s and 41.3% (156 of 378) had velocities of 200–500 nm/s (Fig. 6 B and C). These data suggest that mLnp1 may elicit an effect on nascent junctions by diminishing their mobility, which could facilitate the maintenance or stabilization of nascent junctions.
Discussion
Our prior studies in yeast suggested that Lnp1 plays a role in regulating ring closure. However, the difficulty of imaging the fine structure of the yeast ER network precluded us from testing this proposal. Here we have addressed the role of mLnp1 in the formation and maintenance of the ER network in mammalian cells. As we showed previously for the yeast homolog, mLnp1 is present in the form of distinct puncta that are predominantly localized at three-way junctions (9). Furthermore, as in lnp1∆ yeast, depletion of mLnp1 leads to a collapse of the peripheral ER network into a more compact, sheet-like morphology. The larger dimensions and more open nature of the mammalian peripheral ER network as well as the flat morphology of tissue culture cells has allowed us to follow the dynamics of individual junctions and individual mLnp1 puncta over time in live cells. This analysis has led to several striking findings that provide insights into the function of mLnp1.
By using antibody to localize endogenous mLnp1, we found that only approximately half of all junctions have a detectable concentration of mLnp1. This has allowed direct comparisons of the behavior of mLnp1-positive junctions relative to mLnp1-negative junctions in the same cell that have revealed some important differences. Observation of preexisting junctions shows that mLnp1-positive junctions tend to be less mobile than mLnp1-negative junctions and less likely to undergo junction loss through ring closure. The differences, although significant, are not very large. However, by limiting our observations to nascent junctions formed during the observation period, a very dramatic difference can be documented. A fraction of newly formed junctions go on to acquire mLnp1 protein. Newly formed junctions that do not acquire mLnp1 have a high probability of loss through ring closure and are relatively mobile, whereas those that do acquire mLnp1 have a greatly reduced probability of loss and are less mobile. These results suggest that mLnp1 acts in some way to stabilize newly formed three-way junctions and reduce their mobility. The decreased mobility may in part account for the increased survival probability, as ring closure requires junction movement. As the mLnp1-dependent differences in junction behavior are more striking when observing newly formed junctions in contrast to preformed junctions, one interpretation is that mLnp1 is required initially to stabilize newly formed junctions; however, when mLnp1 has acted on a junction, its continued presence is no longer critical to the survival of that junction. Movement of mLnp1 puncta along tubules away from existing three-way junctions can be observed. Understanding the implications of these observations will require a more detailed understanding of the mechanism of action of mLnp1.
In considering possible mechanisms by which mLnp1 might stabilize three-way junctions, it is important to recall the unique geometry of these structures. Unlike tubules, which exclusively involve positive curvature, three way junctions involve saddle-shaped domains possessing positive curvature in one dimension and negative curvature in an orthogonal dimension. Just as reticulons are thought to add local positive curvature to the ER membrane by acting as a wedge in the bilayer (3, 4), mLnp1 could add local negative curvature by acting as an inverted wedge. Alignment of multiple mLnp1 molecules along the internal angles of a three-way junction could stabilize the structure and also potentially offer some resistance to junction mobility. The observation that mLnp1-GFP puncta can move from one junction to another without obvious changes in brightness suggests that each mLnp1 punctum represents a metastable oligomeric structure. Understanding the nature and composition of these structures and the mechanism by which mLnp1 assembly directs formation of the structure will be important. All members of the Lunapark family contain two predicted transmembrane domains as well as a zinc finger. Protease protection studies indicate that the zinc finger faces the cytoplasmic space (Fig. S7), whereas genetic analysis indicates that it is essential for Lnp1 function in ER morphogenesis (9); however, the exact role of the zinc finger in Lnp1 function is unclear. Although a purely structural role for mLnp1 seems appealing, it is also possible that mLnp1 plays a more transient role, for example, directly or indirectly altering the lipids at the junction to accommodate the complex membrane curvature or serving as a temporary scaffold to direct the assembly of other components. Such a role could explain the more pronounced requirement for mLnp1 in stabilization of nascent junctions relative to preexisting junctions and the observed stability of older junctions that have lost mLnp1.
The apparent requirement for mLnp1 in the stabilization of nascent junctions explains the change in ER architecture observed upon mLnp1 depletion. In the absence of mLnp1, new junctions would continue to be formed; however, they would be much less likely to survive. The increased rate of junction loss through ring closure would lead to collapse of the extended ER network into a more compact, sheet-like structure, as observed upon mLnp1 depletion.
Materials and Methods
Constructs.
mCherry-KDEL and GFP-Atl3 were gifts from Gia Voeltz (University of Colorado at Boulder, Boulder, CO). The construction of human Lnp1-GFP was described previously (9). 3xFLAG-Sec61β was generated by PCR using Sec61β from pAcGFP-Sec61β (gift from Gia Voeltz) as a template, and the PCR product (291 bp) was cloned into the NotI/BamHI sites of p3xFLAG-CMV-7 (Sigma-Aldrich). To construct GST-mLnp1, a BamHI/NotI fragment containing full-length cDNA (1,287 bp) of human Lnp1 was ligated into pGEX-6a-1 (GE Healthcare). His6-mLnp1 was constructed by cloning human Lnp1 cDNA into the NdeI/BamHI sites of pET-15b (Novagen).
Mammalian Cell Culture and Transfection.
COS-7 cells were grown at 37 °C in a CO2 incubator in DMEM with high glucose and 10% (vol/vol) FBS. Transfection of siRNA or plasmid DNA (pSilencer 1.0-U6 shRNA, FLAG-Sec61β, mCherry-KDEL, Lnp1-GFP, and GFP-Atl3) into COS-7 cells was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer’s directions.
RNAi.
The siRNA oligonucleotide (5′-UACGUUGGCACUGGUACGAAU-3′) targeting mLnp1 mRNA was obtained from Qiagen. Two shRNA oligonucleotides specific for mLnp1 mRNA called Lnp1A (5′-GCCTTCAACTGCATATTTA-3′) and Lnp1B (5′-GGGTATGCTTGTGTATCAT-3′) were constructed using the pSilencer 1.0-U6 vector. The siRNA or shRNA vectors were transfected into COS-7 cells using Lipofectamine 2000, and cells were harvested 72 h after transfection. The efficiency of mLnp1 depletion was determined by Western blot analysis.
Immunofluorescence Staining and Confocal Microscopy.
COS-7 cells were seeded on a poly-lysine–coated coverslip grown to 40–70% confluence, washed with PBS solution, and fixed in 4% (wt/vol) paraformaldehyde at room temperature during a 15-min incubation. After the paraformaldehyde was removed, the fixed cells were permeabilized in 0.1% Triton X-100 for 10 min, then treated with 1% BSA in PBS solution plus 0.05% Tween-20 (PBST) for 40–60 min. The primary antibody was added overnight at 4 °C before the cells were washed with 1% BSA in PBST, and incubated with Alexa Fluor 488- or Alexa Fluor 594-conjugated secondary antibodies (Life Technologies) for 1 h at room temperature. The coverslip was mounted on a slide containing Fluoromount-G (Southern Biotech). Fluorescence images were acquired on a Yokogawa spinning disk confocal microscope (Observer Z1; Carl Zeiss)equipped with an electron-multiplying CCD camera (QuantEM 512SC; Photometrics).
To analyze the ER morphology in Fig. 1B, the total area that the ER network covers was determined by outlining the ER at the cell periphery. The area of ER sheets was defined by the Renyi Entrogy threshold setting using ImageJ software (12).
Time-Lapse Imaging of Cortical ER Dynamics.
COS-7 cells were grown in DMEM, transfected with mCherry-KDEL and mLnp1-GFP, and the cells were seeded on a poly-lysine–coated glass bottom culture dish 24 h after transfection. Before the cells were imaged, the culture medium was changed to Opti-MEM medium. Time course images were captured by using a spinning disk confocal microscope as described earlier. The movies were generated by using AxioVision and ImageJ software. The motility of the ER junction or mLnp1 was tracked and measured by using MTrackJ software.
Supplementary Material
Acknowledgments
We thank Gia Voeltz (Department of Molecular, Cellular and Development Biology, University of Colorado at Boulder) for GFP-Atl3 and mCherry-KDEL vectors. This work was supported by the Howard Hughes Medical Institute. Salary support for S.C. and S.F.-N. was provided by the Howard Hughes Medical Institute. P.J.N. was supported by funds from the George E. Palade Endowed Chair.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423026112/-/DCSupplemental.
References
- 1.Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- 2.De Craene JO, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17(7):3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319(5867):1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 5.Shibata Y, et al. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143(5):774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138(3):549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460(7258):978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 8.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Novick P, Ferro-Novick S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat Cell Biol. 2012;14(7):707–716. doi: 10.1038/ncb2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C, Chen LB. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54(1):37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 11.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol. 2013;15(2):169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.