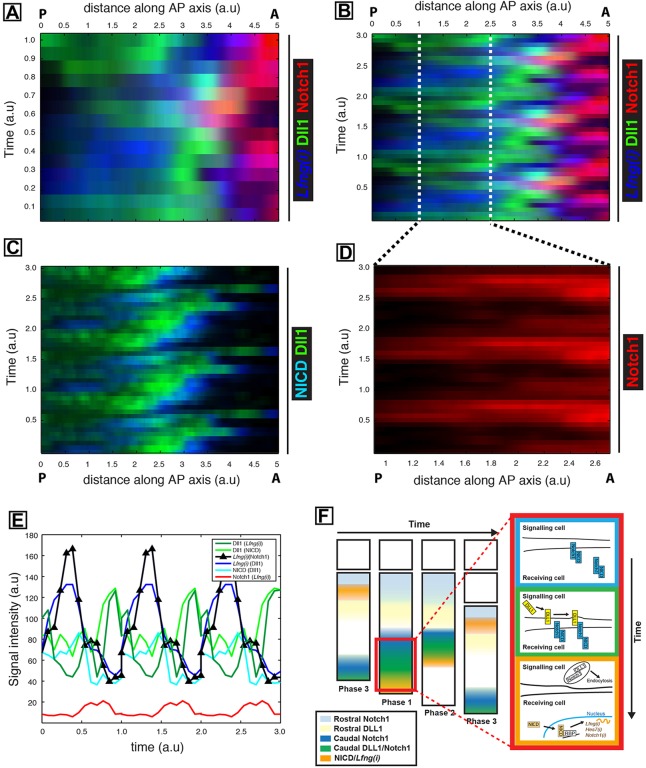
Fig. 7.
Quantification of pulsatile Dll1 and Notch1 protein expression in the caudal PSM of mouse tails. (A) Overlay of kymographs from Fig. 6D [Lfng(i)], Fig. 6E (Dll1) and Fig. 6G (Notch1) reveals their spatiotemporal expression during one oscillation cycle. (B) The periodic extension of the data shown in A highlights the oscillatory nature of the dynamics. (C) The periodic extension of Fig. 6B and E depicts regular oscillations and activity waves of both NICD and Dll1 from caudal to rostral that are out of phase with each other. (D) Kymograph to show the intensity of Notch1 alone within the magnified region from B. (E) The average signal intensity for Notch1, Dll1, activated NICD and Lfng in caudal regions of the periodically extended kymographs is plotted as a function of time. Note that parentheses in the key denote the corresponding paired sample. Hence, for instance, Dll1 appears twice, as it is paired with both NICD and Lfng(i). a.u. arbitrary units. (F) A proposed model. Pulses of Notch1 protein followed closely by Dll1 originate in the caudal PSM, which initiates a sequence of events detected as spatially separated events along the PSM, depicted as coloured bands of expression. The red boxed region that is magnified to the right contains coloured boxes to show the details of events in each colour band in the PSM. Blue, Notch is translated and translocated to cell membranes; green, Notch 1 and Dll1 are translated and translocate to cell membranes, leading to transactivation; orange, transactivation leads to cleavage and release of NICD, which translocates to the nucleus and activates downstream transcription of Notch target genes (including Notch1, Lfng and Hes7), while Dll1 and/or extracellular Notch1 are endocytosed to be recycled or degraded. Phase 1, 2 and 3 refer to well-established phases of Lfng/NICD expression domains.