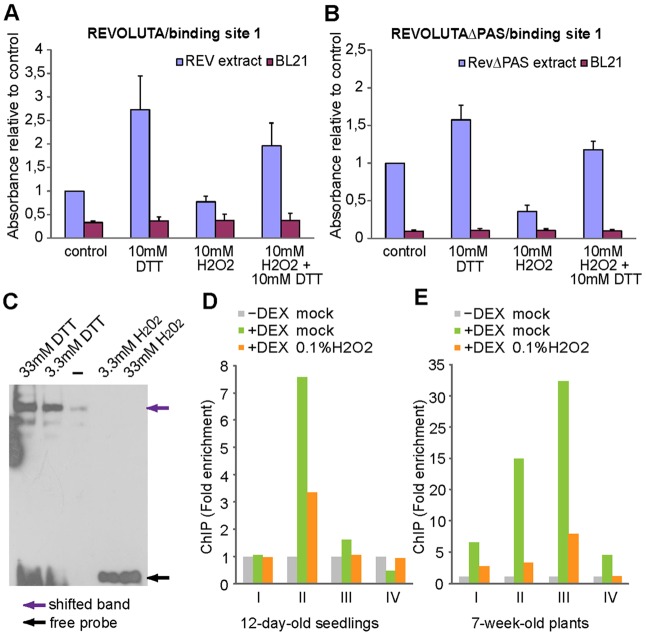
Fig. 4.
Redox-mediated regulation of REVOLUTA-DNA-binding capability and influence of the PAS domain. (A,B) Redox-DPI-ELISAs. The DNA-protein interaction assays were performed by using 5′ biotinylated complementary annealed oligonucleotides coupled to a streptavidin-coated ELISA plate. Crude E. coli extracts (25 µg) expressing recombinant REV or REVΔPAS were pre-incubated with different concentrations of DTT and H2O2 to examine a redox state-dependent binding of REV. In order to test the reversibility of the redox effect, high concentrations of H2O2 were added first and then oxidizing conditions were reversed by addition of DTT. After binding, biotinylated DNA-protein complexes were detected using anti His-HRP conjugated antibodies. Results for REV binding site 1 of the WRKY53 promoter are shown. E. coli BL21 cells transformed with the empty vector were used as background control. (C) Non-radioactive electrophoretic mobility shift assays. Purified GST-REV protein was incubated with a biotinylated oligonucleotide containing the HB9-binding motif (Wenkel et al., 2007) in the presence of different redox conditions. After gel electrophoresis and subsequent blotting, the biotinylated DNA probe was detected with a HRP-strepatividin substrate. (D,E) Chromatin-immunoprecipitation qPCR assays of 35S::FLAG-GR-REVd plants. Twelve-day-old seedlings (D) and 7-week-old transgenic plants (E) were treated with mock substrate (0.1% ethanol), DEX or 0.1% H2O2 and DEX. H2O2 was given 15 min prior to 45 min of DEX induction. Fold enrichment for the same primer sets as in Fig. 1 is shown.