Figure 1. Overview of the MinSpan algorithm.
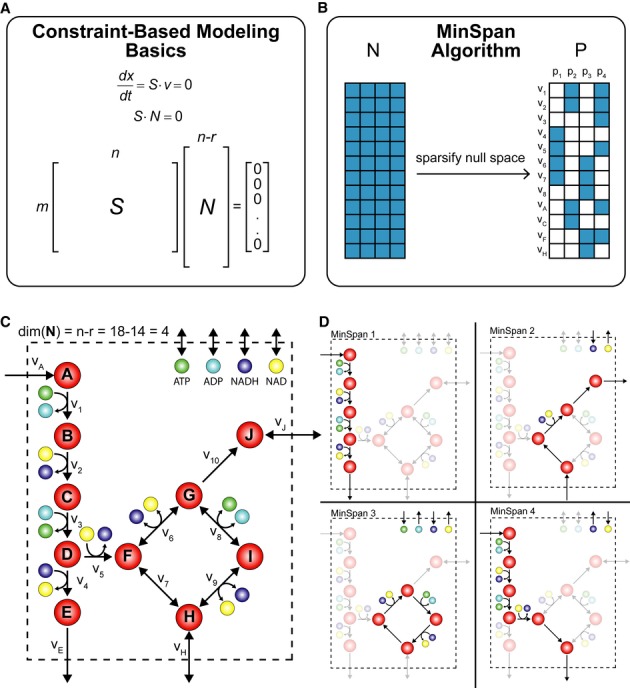
- A metabolic network is mathematically represented as a stoichiometric matrix (S). Reactions fluxes (v) are determined assuming steady state. All potential flux states lie in the null space (N).
- The MinSpan algorithm determines the shortest, independent pathways of the metabolic network by decomposing the null space of the stoichiometric matrix to form the sparsest basis.
- A simplified model for glycolysis and TCA cycle is presented with 14 metabolites, 18 reactions, and a 4-dimensional null space. Reversible reactions are shown.
- The four pathways calculated by MinSpan for the simplified model are presented, two of which recapitulate glycolysis and the TCA cycle, while the other two represent other possible metabolic pathways. The flux directions of a pathway through reversible reactions are shown as irreversible reactions.
