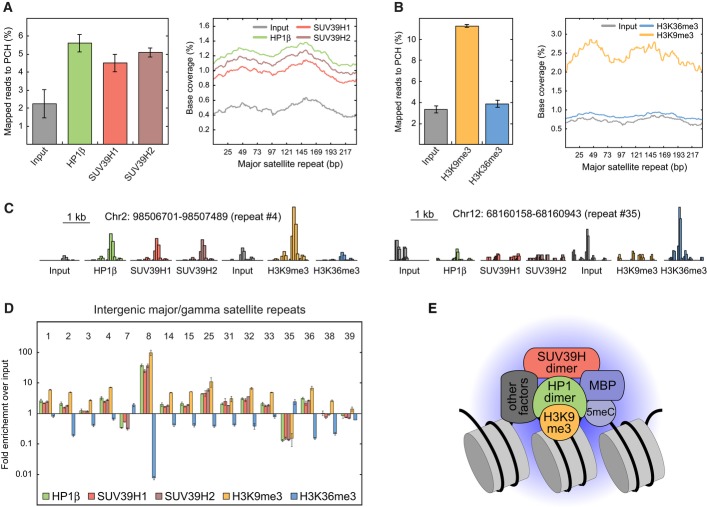
Figure 4. Genome-wide ChIP-seq analysis of HP1, SUV39H, and H3K9me3 enrichment at major satellite repeats.
- Fraction of total sequencing reads that mapped to the consensus sequence of major satellite repeats (gamma satellites, GSAT) after ChIP-seq of HP1β, SUV39H1, or SUV39H2. Mild sonication conditions were applied in neural progenitor cells (NPCs). The histogram shows the distribution of the mapped reads along the consensus sequence. Error bars correspond to SD.
- Same as panel A but for ChIP-seq of H3K9me3 and H3K36me3 and with stronger sonication conditions.
- Representative sequencing read distributions at interspersed major satellites that can be uniquely identified in the genome. Two repeats located on chromosomes 2 and 12 (no. 4 and no. 35 in panel D) are depicted. HP1 and SUV39H were enriched at the transcriptionally inactive repeat (no. 4) that additionally had a high level of the repressive H3K9me3 mark and a low level of the activating H3K36 trimethylation. At the actively transcribed repeat (no. 35) marked by H3K36me3, binding of SUV39H and HP1 was at background levels. The height of the histogram is normalized to total reads.
- Enrichment of all mapped sequencing reads to interspersed GSAT regions over input control. The 16 regions that can be uniquely mapped are shown. Error bars correspond to SD.
- Composition of the HP1-SUV39H nucleation complex as inferred from the enrichment of stably bound protein in PCH, protein–protein interaction measurements and ChIP-seq analysis. The postulated complex comprises a HP1 dimer binding H3K9me3-modified nucleosomes and interacting with a SUV39H dimer. Binding of SUV39H is further stabilized by methyl-binding proteins (MBPs) that recognize 5meC and possibly by other interacting factors.