Figure 7. Quantitative model for H3K9 trimethylation in PCH.
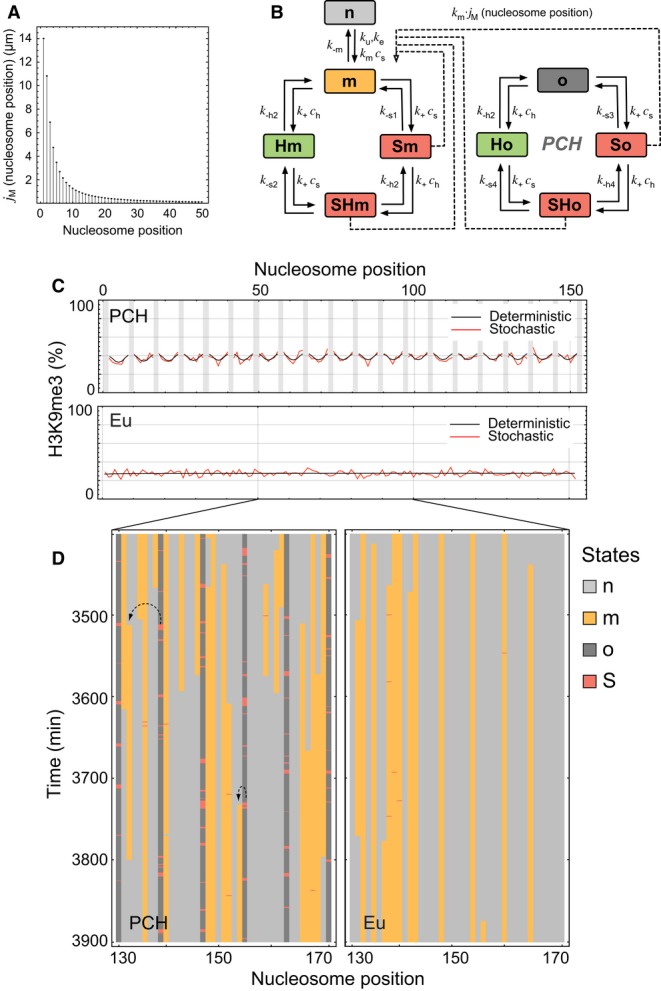
- Probability of interactions between two nucleosomes due to chromatin looping as expressed by their local molar concentration jM. Based on the data for chromatin interactions during recombination in living cells (Ringrose et al, 1999), the dependence of jM on the separation distance from a SUV39H-bound nucleosome was calculated as described previously (Rippe, 2001; Erdel et al, 2013).
- PCH network model scheme. The model includes transitions between the following states according to the indicated rate constants: “n”, nucleosome without H3K9me3; “m”, H3K9me3 modified; “o”, origin that represents a high-affinity binding site for SUV39H. HP1 and SUV39H bound to these sites are represented by H and S, respectively, to form the states “Hm”, “Sm”, “SHm”, “Ho”, “So”, and “SHo”. All SUV39H-bound states (red) enhance methylation of adjacent unmodified nucleosomes through chromatin looping. See text and Supplementary Table S7 for further details.
- Steady-state distribution of methylation levels for a chromatin segment in PCH or euchromatin. The deterministic and stochastic distributions are depicted in black and red, respectively. Stochastic solutions were averaged over 5,000 min. The nucleation origins (both occupied and unoccupied) are marked in gray.
- Stochastic simulation for a region of 40 nucleosomes. Same color code for nucleosome states and origins as in panel B.
