Figure 1. Analysis of TIS sequences via FACS-seq.
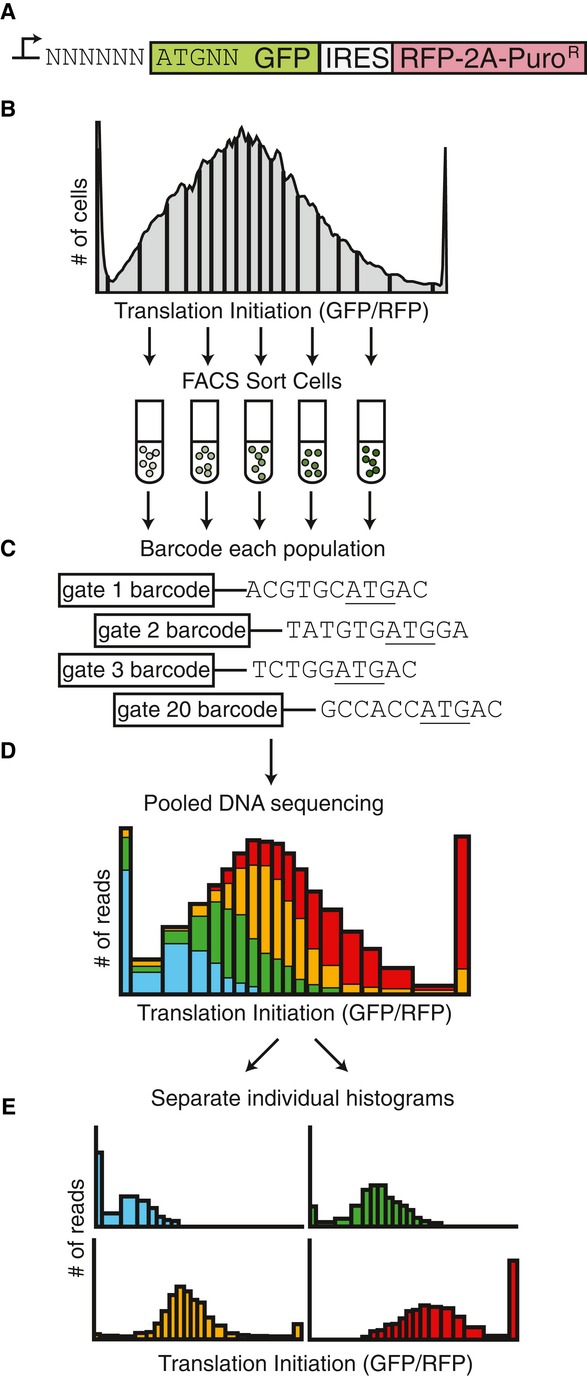
- A TIS reporter library was created by degenerate PCR to create a random permutation of the six bases upstream and two bases downstream of a GFP start codon, resulting in a library size of 65,536 TIS sequences. An IRES followed by RFP was used to normalize the GFP expression.
- Stably transduced cells were sorted based on translation efficiency Analysis of TIS sequences via FACS-seq (GFP/RFP). 20 gates were drawn such that each gate contained 5% of the total library population. Cells that were off-scale were collected in the last gate.
- The TIS sequences from each sorted population were PCR amplified and barcoded.
- The barcoded TIS library was pooled and sequenced. The number of reads for each TIS sequence and barcode combination was counted.
- An individual histogram for each TIS sequence is created in silico.
