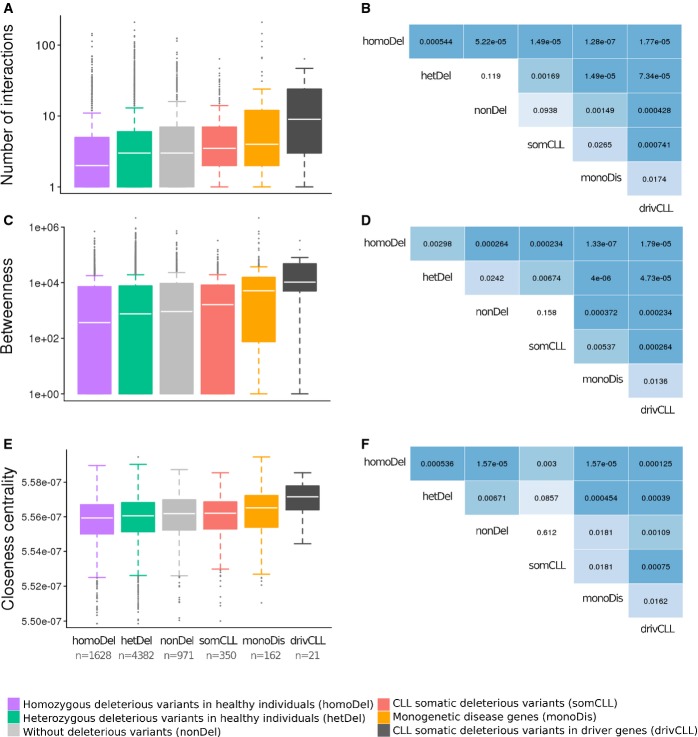
Figure 3. Connection degree, betweenness and closeness centrality of proteins affected by deleterious variants.
- From left to right: Number of interactions in proteins affected by deleterious variants in both alleles (homozygosis), in only one allele (heterozygosis), not affected by any deleterious variant, proteins affected (in homozygosis or heterozygosis) in a pathological condition (somatic variants in CLL), proteins affected by monogenic diseases (listed in Supplementary Table S2) and the subset of somatic variants in CLL which occur in cancer driver proteins (Vogelstein et al, 2013) (listed in Supplementary Table S3).
- Significance of the comparisons tested by the rank sum (Mann–Whitney U-test) with FDR multiple testing adjustments.
- Betweenness in the same groups of proteins as in (A).
- Significance of the comparisons tested as in (B).
- Closeness centrality in the same groups of proteins as in (A).
- Significance of the comparisons tested as in (B).
Data information: In this boxplot representation boxes correspond to 50% of the observed values and error bars to 90%.