Fig. 7. DNase I footprinting analysis of PdxR binding to the mutant pdxRS regulatory regions.
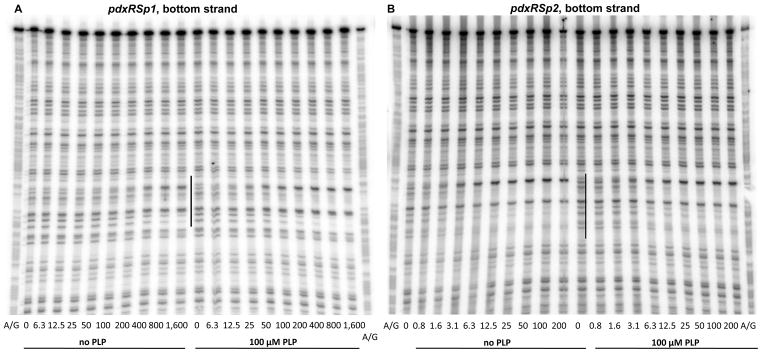
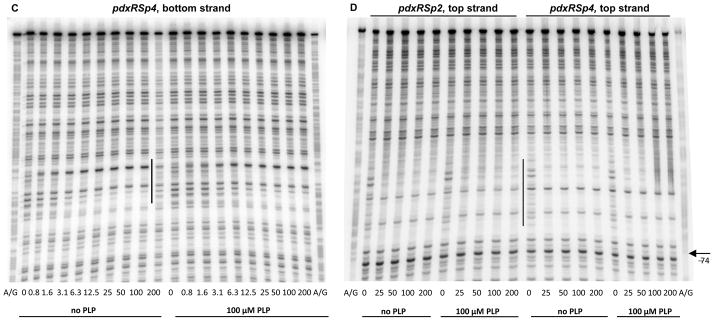
pdxRSp1 (A), pdxRSp2 (B, D), and pdxRSp4 (C, D) DNA fragments, radioactively labeled on the bottom (A, B, C) or top (D) strand, were incubated with increasing concentrations of purified PdxR and in the absence or presence of 100 μM PLP. PdxR monomer concentrations used (nM) are indicated below each lane. The corresponding A + G sequencing ladder is shown in the left and right lanes. The protected areas are indicated by vertical lines.
