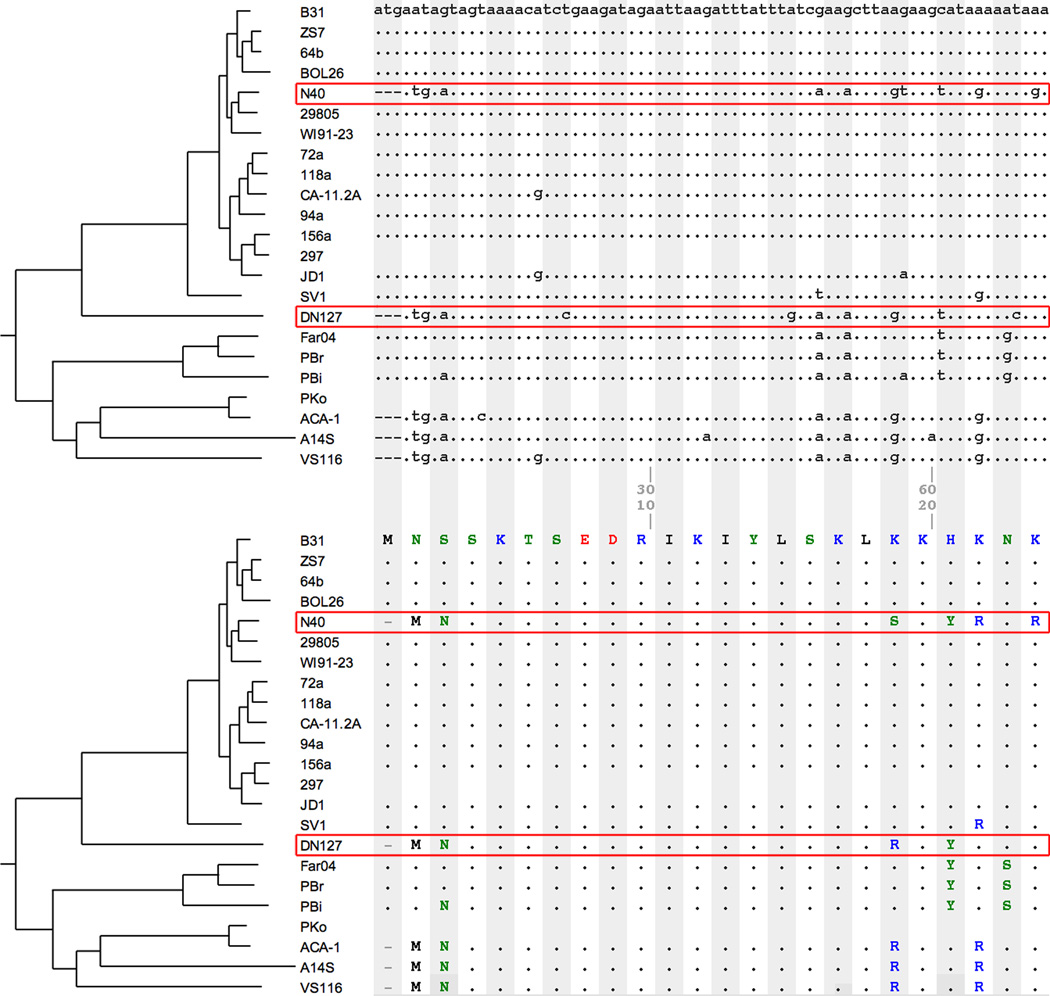
Figure 6. Genomic phylogeography: A cross-species hybridization.
(Top Panel) A codon alignment of the 5’ sequences at the main chromosomal locus 0082 (encoding an uncharacterized conserved protein). B31 sequence is used as the reference and an “.” in other sequences represents the same nucleotide as in the B31 sequence at the same aligned site. Neighboring codons are shaded in alternating colors. The corresponding amino-acid alignment (Bottom Panel) shows coding consequences (synonymous or non-synonymous) of individual nucleotide variations. The N40 sequence at this region displays a large phylogenetic inconsistency: it is more similar to the sequences of non-B. burgdorferi s.s. orthologs than to its con-specific strains. Since all outgroups of B. burgdorferi s.s. used here are distributed exclusively in Europe or Western US, it is likely that the donor is a DN127-like species in Eastern US, such as B. kurtenbachii (Margos et al., 2010). Further analysis identified the two ends of this single cross-species gene-conversion event and showed that it affects a 2.2-kilobase region from the 3’ end of the 0081 locus (encoding a permease) to the 5’ end of the 0084 locus (encoding an aminotransferase). While this gene-conversion event is not obviously adaptive, this analysis shows that (i) there is cross-species hybridization in Northeast USA, (ii) the hybridization may be recent (see Section 7), and (iii) evolutionary genomic analysis is powerful to reveal population history. In this case, signatures of recombination are discovered based on phylogenetically mismatched SNPs (“homoplasies”). The graph was prepared by using BorreliaBase.org, a phylogeny-centered browser designed for evolution-informed comparative analysis of Borrelia genomes (Di et al., 2014). Future studies will screen for genome-wide signatures of species hybridization and population admixture.