Figure 1.
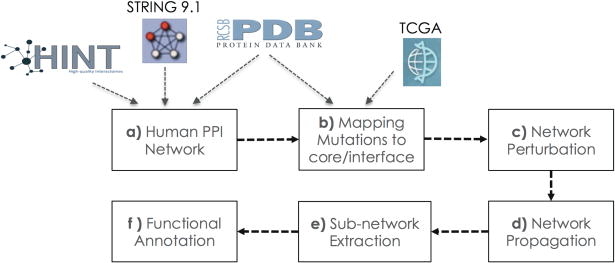
A pipeline for mining molecular cancer related sub-networks accounting for different effects of distinct mutations. Steps include network assembly (a), mapping of mutations to interface versus core residues of cancer genes (b), removal of affected edges (c), extraction of associated protein sets (d & e) and functional annotation (f).
