Fig. 4.11.
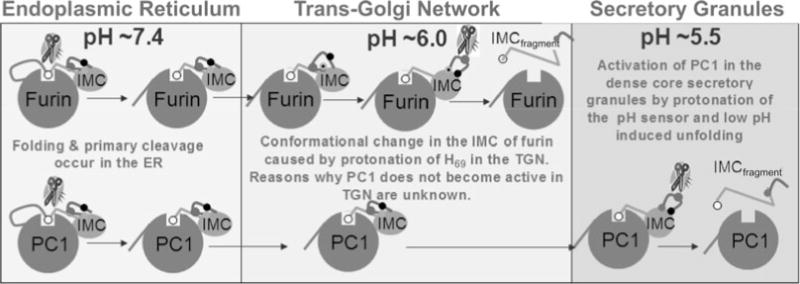
A cartoon model depicting putative mechanism for activation of furin and PC1/PC3 through protonation of H69. Both furin and PC1/PC3 undergo their first cleavage at a pair of conserved dibasic residues. This cleavage allows the non-covalent complexes of furin and PC1/PC3 to exit the ER into the TGN. Here, His69, the pH sensor in furin, gets protonated and induces a conformational change within the pH-sensitive loop. This exposes the internal cleavage site at Arg75, which can undergo proteolysis that releases inhibition. Although the residue corresponding to His69 in furin is conserved, PC1/PC3 does not undergo the second internal cleavage and suggests that additional residues must be involved in fine-tuning the pH sensitivity of individual PCs. Upon internal cleavage the inhibitory C-terminal region of the IMC is released and renders the catalytic site free for proteolysis.
