Fig. 2.
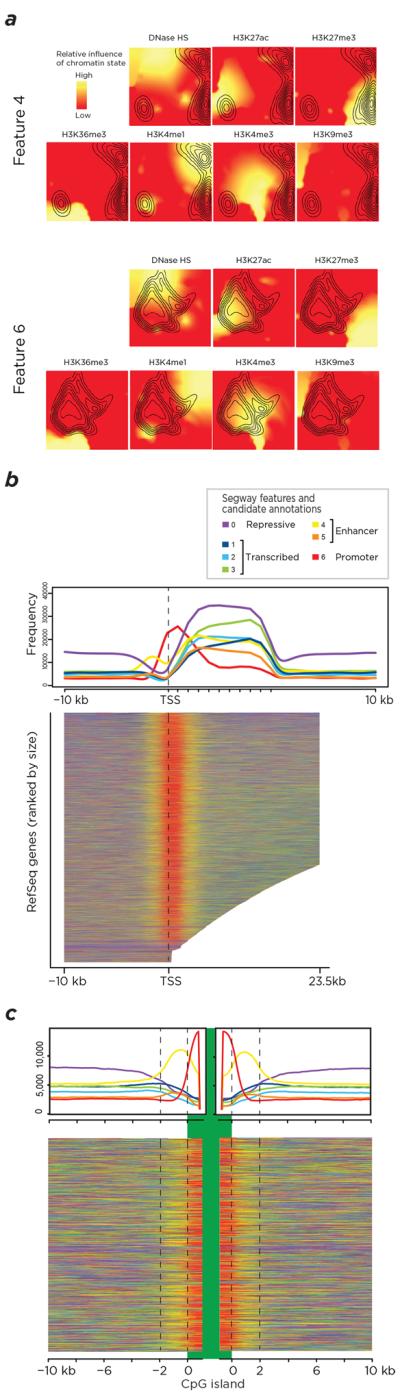
Empirical annotation of the CD34+ HSPC genome based on chromatin features reveals candidate cis-regulatory element locations. Panel (a) shows a contour plot of the regions within the self-organizing map (SOM) where Segway features 4 (above) and 6 (below) enrich, showing feature 4 to be composed of loci where H3K4me1 and H3K27me3 occur, while the loci composing feature 6 contain the H3K4me3 and H3K27ac modifications. Consistent with these findings, panel (b) shows feature 6 (red) to be enriched at the transcription start site for a metaplot (top) and a heat map (below) of all RefSeq genes, indicating promoter characteristics, while feature 4 (yellow) flanks this region and is consistent with enhancers in a poised state. In panel (c), similar metaplot (top) and heat map (below) representations of the 2 kb flanking CpG islands demonstrate strong enrichment in feature 4, indicating that these “CpG island shores” in fact represent candidate enhancers in this cell type.
