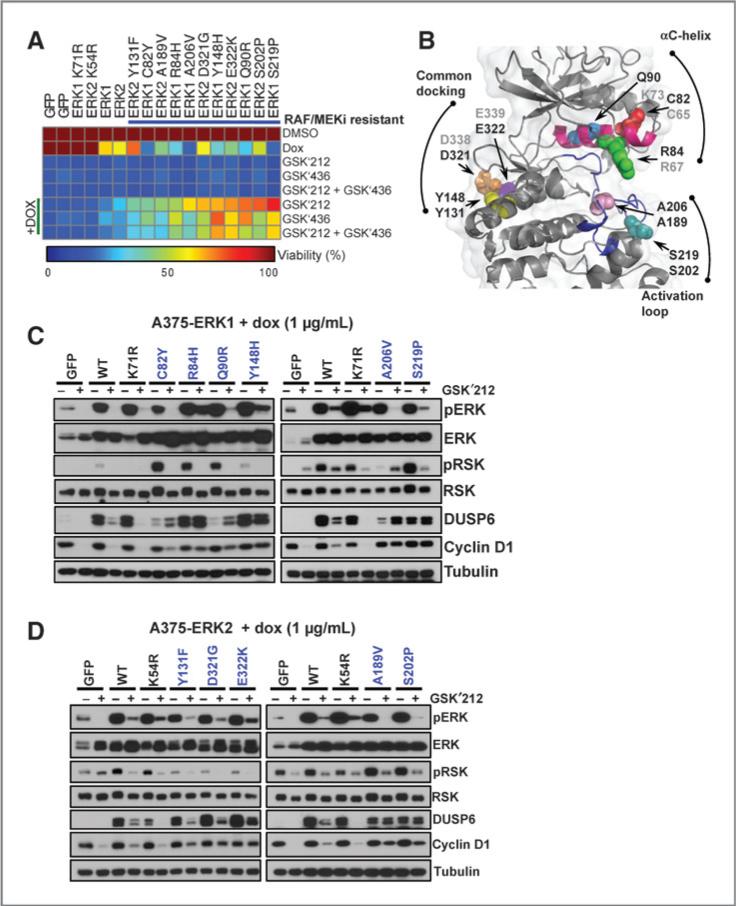
Figure 4.
Pharmacologic and biochemical validation of ERK mutations that confer resistance to RAF/MEK inhibitors. A, A375 cells expressing tet-inducible RAF/MEK inhibitor resistance mutations were analyzed for viability in the presence of trametinib (GSK′212), dabrafenib (GSK′436), or trametinib + dabrafenib (GSK′212 + GSK′436), with or without DOX. Cell viability was normalized to DMSO, and values are depicted in the heat map. Alleles are sorted by sensitivity to trametinib + DOX. GFP, wild-type ERK1/2, and kinase-dead ERK1K71R and ERK2K54R served as controls. B, structural localization of validated RAF/MEK inhibitor–resistant alleles (spheres) mapped onto the ERK2 crystal structure (cartoon; PDB: 2ERK). The αC-helix (magenta) and the activation loop (blue) are labeled. The nonvalidating/ untested analogous ERK1/2 alleles are labeled in gray. C and D, cells expressing validated ERK1 (C) or ERK2 (D) RAF/MEK inhibitor resistance alleles were exposed to DOX with or without trametinib and pERK, pRSK, DUSP6, and cyclin D1 expression was analyzed by immunoblotting.