Figure 1. Organization and alternative splicing (circled a–n) of the killifish esr1 gene, as determined by targeted PCR cloning and sequence analysis of ERα cDNAs.
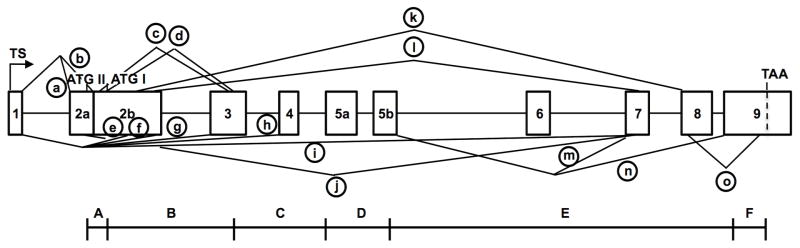
See text for the cloning strategy, Table 1 for primer sequences and Table 2 for mRNA characteristics and GenBank Accession numbers. Cloned cDNAs were assembled and mapped onto the marine medaka genome (Ensembl.org, #ENSORLG00000014514) and subsequently confirmed by mapping onto a preliminary draft of the killifish genome (Fundulus.org) and by PCR cloning and sequencing of selected introns. See Table S1 for comparison of exon and intron sizes in killifish, medaka and zebrafish. A single transcription start site (TS) and two translation start sites (ATGI, ATGII) were identified. A transcript with alternative splicing between exon 1 and an internal acceptor site in exon 2 (circled a) resulted in a short transcript lacking exon 2a and predicting translation from ATGI in exon 2b. A second transcript with alternative splicing between exon 1 and exon 2a (circled b) is predicted to begin translation at ATGII in exon 2a. Additional alternative splicing events (circled c – m) resulted in mRNAs with deletions of one or more internal coding exons. The previously cloned ERαx mRNA splice variant (circled n) is included for reference (Greytak and Callard, 2007). The six functional domains (A–F) of the human ERα protein (Krust et al., 1986) are shown below the corresponding exons. Although the esr1/ESR1 gene has 8 coding exons in all vertebrates, by convention the first coding exon is termed exon 2 in zebrafish and rainbow trout (Menuet et al., 2004; Pakdel et al., 2000) but exon 1 in mouse and human (Ponglikitmongkol et al., 1988; White et al., 1987).
