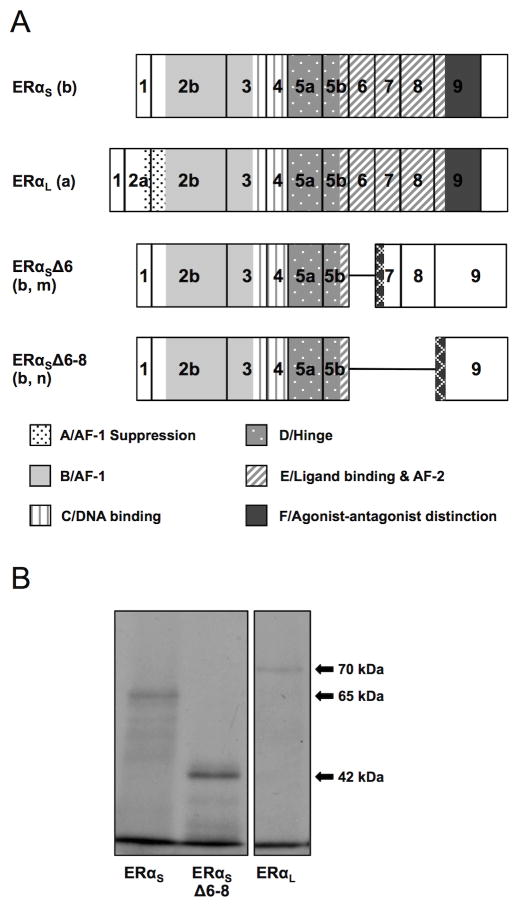
Figure 2. (A) ERαS, ERαL ERαSΔ6 and ERαSΔ6–8 isoform structure as predicted in silico. (B) Short, long and Δ6–8 ERα isoform size as confirmed by in vitro transcription/translation.
(A). Lower case letters in parenthesis are keyed to the splicing events shown in Figure 1. The previously identified ERαS was produced from the splicing of exon 1 to exon 2b (b), and was predicted to use ATGI resulting in a protein of 573 aa lacking the A domain. The newly identified ERαL was produced from the splicing of exon 1 to exon 2a (a), and its translation is predicted from ATGII in exon 2a resulting in a protein of 620 aa containing all six functional domains. A deletion variant lacking exon 6 (Δ6, m) was paired with both the short and long 5′ends predicting a 333 (short) or 380 (long) aa protein, respectively (only the short is shown). A deletion variant lacking exons 6, 7, and 8 (Δ6–8, n) was paired with both the short and long 5′ends, predicting a 334 (short) or 381 (long) aa protein, respectively (only the short is shown, b). Both Δ6 and Δ6–8 are predicted to be missing most of the E and all of the F domains. The key to the functional domains of the ERα protein as defined in human (Krust et al., 1986) and rainbow trout (Pakdel et al., 2000) is shown at the bottom of the panel. Hatched regions indicate novel amino acid sequences generated by frame shifts. Predicted protein structure of additional cloned mRNAs is included in Supplementary materials (Figure S1). Characteristics of the variant mRNAs and predicted proteins are summarized in Table 2.
(B). In vitro translation of ERαS, ERαL, and ERαSΔ6–8 with TnT-coupled reticulocyte lysate in the presence of 35S-labeled methionine resulted in proteins of 65, 70, and 42 kDa, respectively, corresponding with the in silico size predictions.