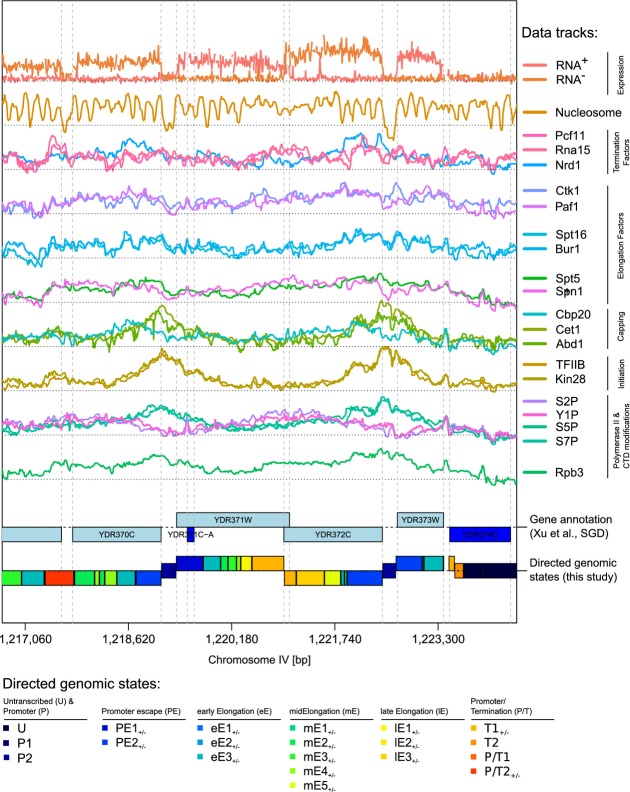
Figure 2. De novo annotation of directed genomic states from genomewide transcription data in yeast using bdHMM.
Inputs for the bdHMM are the following, from top to bottom: strand-specific wild-type RNA levels, occupancy maps of nucleosomes, 3 termination factors, 6 elongation factors, 3 capping factors, 2 initiation factors, 4 CTD modifcations and 1 core Pol II member (Rpb3). Inferred directed genomic states are shown as colored boxes in the lowest track (see color legend beneath) where expressed states on the + (respectively −) strand are positioned above (respectively under) the axis, and not expressed states are centered on the axis. Previous transcriptome annotation is shown in the 2nd track from the bottom.