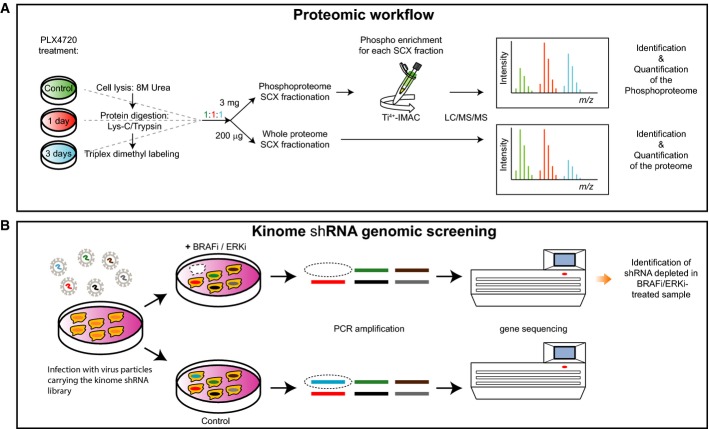
Figure 1. Proteomic and genomic workflows.
- Cell lysates from control samples and samples derived from 1 and 3 days after PLX4720 treatment were digested with Lys-C/Trypsin, labeled by triplex dimethyl approach and mixed in 1:1:1 ratios. For protein expression analysis, 200 μg of digested lysate was fractionated by SCX and each fraction was analyzed by LC/MS/MS to determine the relative protein expression levels for every time point compared to the control. For the phosphoproteome, 3 mg of digested lysate was fractionated by SCX and each fraction was enriched for phosphopeptides by Ti4+-IMAC prior to LC/MS/MS analysis.
- Melanoma cells were transduced with a lentiviral kinome library, containing ˜4,000 shRNAs targeting ˜500 kinases. Cells were treated either with DMSO (control) or with BRAFi or ERKi. Genomic DNA was isolated, and hairpins were amplified by PCR. Using deep sequencing, the hairpins that specifically dropped out in the treated sample were identified. In this case, the absence of the blue bar in deep sequencing indicates schematically a synthetic lethal effect of the shRNA and BRAFi/ERKi.