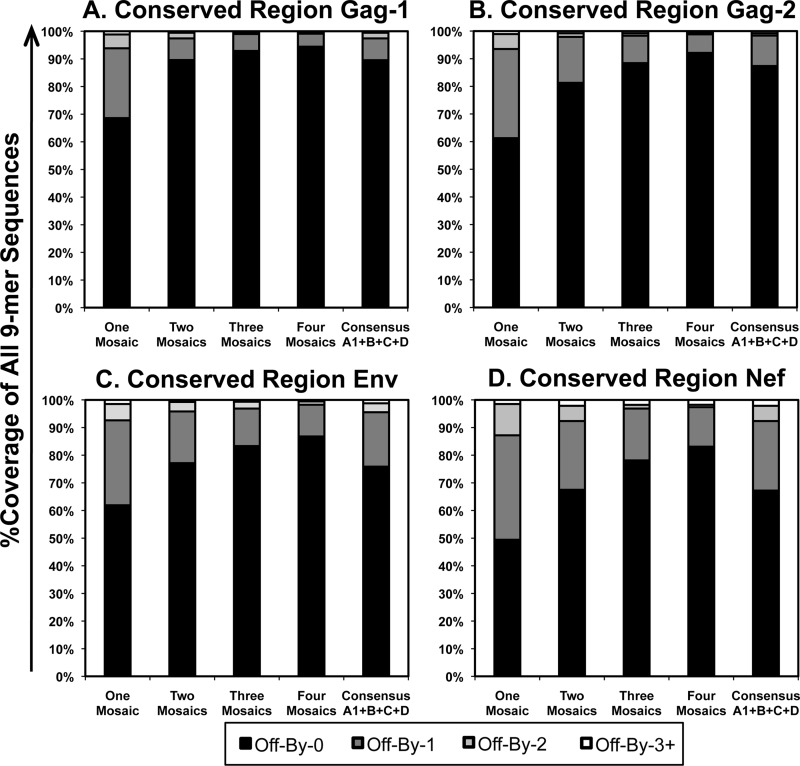
FIG 1.
Coverage of selected conserved regions of the HIV-1 proteome by mosaic and clade consensus sequences. Mosaic sequences optimized to cover HIV-1 M-group Gag, Env, and Nef 9-mer stretches were assessed for coverage of all 9-mers in comparison to a combination of clades A1, B, C, and D for the four conserved regions. (A) Gag-1 (Gag amino acids 148 to 214); (B) Gag-2 (Gag amino acids 250 to 335); (C) Env (Env amino acids 521 to 606); (D) Nef (Nef amino acids 106 to 148). “Off-By-0” refers to the percentage of all possible 9-mer sequences within the Los Alamos HIV Database curated M-group sequences that were perfectly represented within the mosaic sequence(s), “Off-By-1” refers to the percentage that were represented by a one-amino-acid mismatch within the mosaic sequence(s), etc.