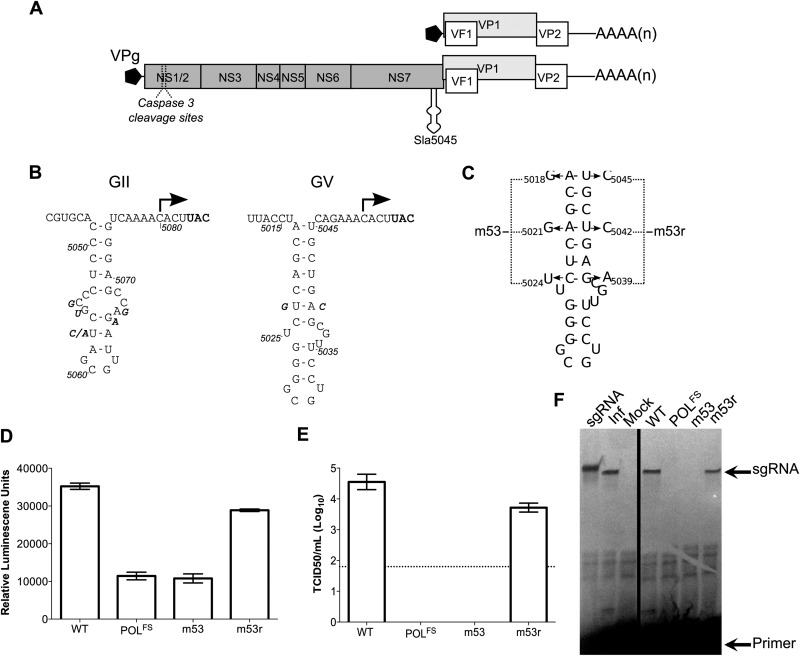
FIG 1.
The stability of Sla5045 is required for norovirus infectivity, replication, and sgRNA synthesis. (A) A schematic of the murine norovirus (MNV) genome, highlighting the four open reading frames and Sla5045. (B) Genetic conservation of the Sla5045 sequences on the norovirus negative-sense viral RNA. The position of the potential norovirus subgenomic RNA initiation site is indicated with an arrow. The position of the VP1 initiation codon on the positive-sense RNA is highlighted in bold, and sequence variation observed in different isolates is shown as bold italic. For genogroup II (GII) noroviruses, the sequence of the GII isolate Hu/GII.4/MD-2004/2004/US is shown (DQ658413), with variation between the following isolates shown: FJ595907.1, AB447425.1, HM635151, HM635164, JF262610.1, and JF262592. For genogroup V (GV) murine norovirus, the sequence of the CW1 isolate (DQ285629) is shown, with variation between the following isolates displayed: JN975491 and JN975492. Note that for simplicity, only sequences showing variation in the stem-loop sequence were used in the analysis and shown in the figure. (C) Structure of Sla5045 and the mutations introduced to generate the mutants m53 and m53r as described previously (16). In the case of m53r, the mutations shown are in addition to those present in m53. (D) Luciferase replicon-based analysis of the effect of Sla5045 disruption. Luciferase levels represent the mean for triplicate independent samples. (E) Virus yield after reverse genetics recovery of full-length cDNA constructs containing either wild-type (WT) MNV, a polymerase frameshift in the NS7 (POLFS), m53, or m53r. The virus titers represent the mean values obtained at 24 h posttransfection of viral cDNA expression constructs. The detection limit of the assay is indicated with a dotted line. In all cases where shown, error bars represent the standard error of the mean (SEM). (F) Primer extension-mediated detection of sgRNA synthesis in MNV-infected cells (Inf) or cells transfected with capped RNAs of WT, POLFS, m53, or m53r clones. In vitro-transcribed sgRNA was used as a positive control (sgRNA) and produces a product 3 nt longer due to the addition of three 5′ G nucleotides by T7 RNA polymerase. Note that 1,000-fold less RNA was used for the reaction using RNA from infected cells.