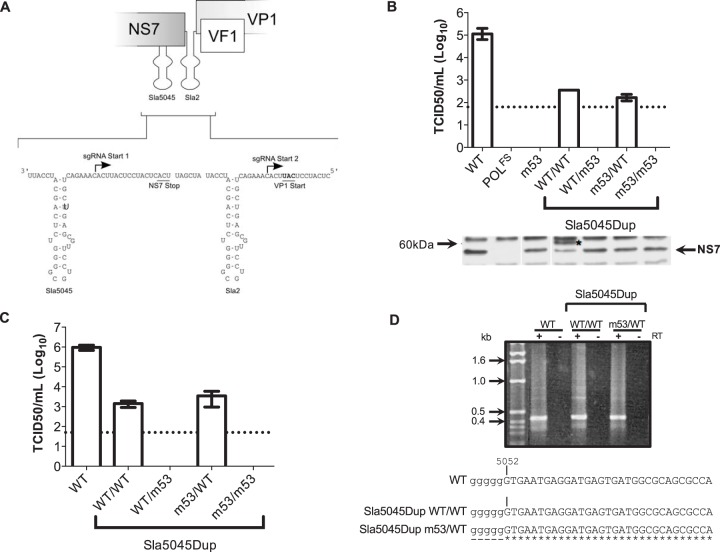
FIG 5.
Duplication of Sla5045 enables mutagenesis in a noncoding context. (A) Schematic illustration of the reconstruction of Sla5045Dup stem-loop duplication constructs. The position of the authentic Sla5045 on the antisense RNA genome is shown, and the introduced additional Sla5045 is shown as Sla2. For simplicity, only wild-type copies of the stem-loop are shown. The positions of the NS7 stop and VP1 start codons on the corresponding positive-sense viral RNA are also shown for reference. (B) Virus yield following reverse genetics rescue of capped in vitro-transcribed RNA of wild type (WT), polymerase frameshift (POLFS), m53, or Sla5045 stem-loop duplication (Sla5045Dup) constructs. The Sla5045Dup constructs are shown using the nomenclature copy1/copy2 as described in the text. Reverse genetics recovery was performed as described in Materials and Methods using in vitro-transcribed, enzymatically capped RNA and the virus yield at 24 h posttransfection determined by TCID50. A Western blot analysis for NS7 expression is shown below the virus yield data. The asterisk indicates a greater-than-full-length NS7 product observed in cells transfected with SLa5045Dup WT/WT. (C) Virus yield assay following a single passage of virus recovered following RNA transfection of wild-type MNV (WT) or various Sla5045 stem-loop duplications. The dotted line represents the detection limit of the assay, with error bars representing the SEM. (D) 5′ RACE analysis of WT MNV or Sla5045 stem-loop duplications. Viral RNA was isolated from infected cells and subjected to 5′ RACE analysis. Samples were prepared with or without reverse transcription (RT) and then subjected to PCR as described in Materials and Methods. The wild-type MNV virus was included as a control. Samples were then sequenced and aligned as shown. The lowercase poly(G) tract is introduced as a result of the 5′ RACE methodology.