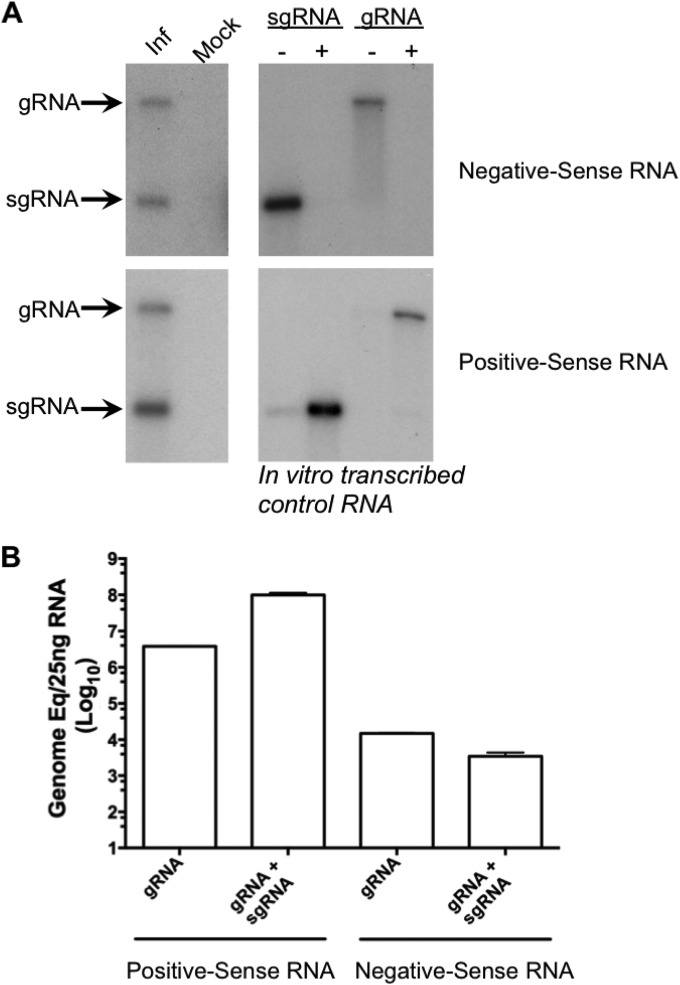
FIG 8.
Negative-sense subgenomic RNA is produced during MNV replication. (A) Northern blot analysis of RNA isolated from either mock-infected (mock) or MNV-infected (Inf) cells. A 1.5-μg portion of RNA harvested 10 h postinfection of cells with a multiplicity of infection of 5 TCID50 per cell was denatured by glyoxylation prior to agarose gel electrophoresis. Positive- and negative-sense RNAs were detected using strand-specific RNA probes as described in Materials and Methods. Control in vitro-transcribed RNAs representing the positive-sense (+) or negative-sense (−) viral RNAs were used as a control. (B). Quantitative real-time PCR analysis of the RNA sample shown in panel A. The RNA was analyzed using a strand-specific RT-qPCR assay designed to detect only the genomic RNA or both genomic and subgenomic RNAs simultaneously. The genome copy number is shown as genome equivalents per 25 ng of total RNA and was determined by comparison to in vitro-transcribed control RNAs. The data shown represent the mean and standard deviation from 4 independent repeats.