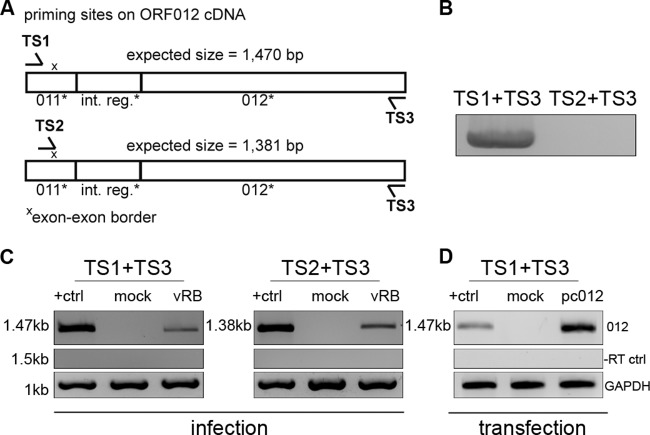
FIG 2.
Analysis of ORF012 splicing in MDV infection by RT-PCR. (A) Positions of primer binding regions on ORF012 cDNA. Two sets of primers specific for the 5′ and 3′ coding regions of the ORF012 transcript (TS1 and TS3) or the exon-exon border (TS2) were used. The primer sequences are listed in Table 1. (B) Primers were tested using genomic RB-1B DNA. Note that no product should be obtained with the primer combination TS2 and TS3 due to the absence of the exon-exon border in genomic DNA. (C) Total RNA of MDV-infected or mock-infected CEC was extracted 5 days p.i. The RNA was reverse transcribed into cDNA with the indicated primers. Amplification products obtained by subsequent PCR were analyzed by agarose gel electrophoresis. cDNA prepared from DF-1 cells transfected with an expression plasmid encoding an intronless ORF012 construct (termed pc_012Δint) served as a positive control and size marker (lanes +ctrl). The amplicons were subjected to Sanger sequencing and showed 100% identity with the predicted mRNA. cDNA of chicken GAPDH mRNA served as an internal control (bottom), and RT-negative control reactions excluded genomic DNA contamination (middle). (D) To show that splicing of ORF012 is independent of other viral factors, total RNA of DF-1 cells transfected with the pc_012Δint, pc012, or pcDNA3.1 vector (negative control) was extracted 24 h after transfection. Samples were subsequently treated as described for panel C. Sequencing revealed 100% identity with the predicted ORF012 mRNA sequence.