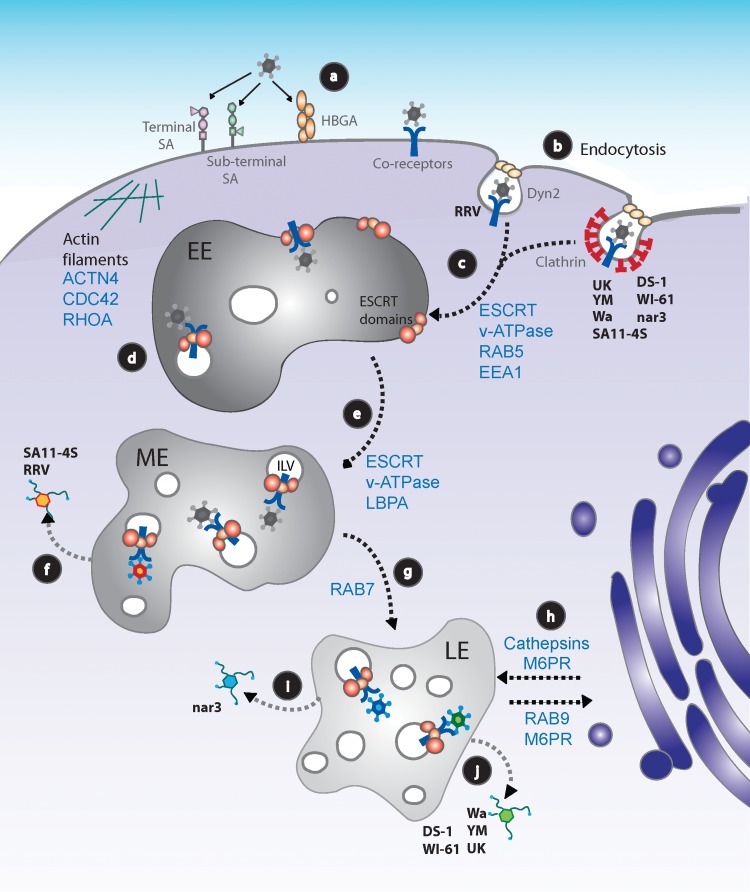
FIG 1.
Working model for rotavirus cell entry into MA104 cells. (a) RVs attach to the cell surface through different glycans, depending on the virus strain. After initial binding, the virus interacts with several coreceptors concentrated at lipid rafts. All known coreceptors are represented as a single blue Y symbol. (b) RVs are internalized into cells by clathrin-dependent or -independent endocytic pathways, depending on the virus strain. (c) Regardless of the endocytic pathway used, all RV strains reach EEs in a process that depends on RAB5, EEA1, and probably on HRS and the vacuolar ATPase (v-ATPase) (11). (d) At the EE, the virus probably begins to be internalized into the endosomal lumen through the action of VPS4A. (e) EEs progress to MEs, with a progressive decrease in pH and intraendosomal calcium concentration through the function of the v-ATPase; during this process the formation of ILVs increases. (f) E-P rotaviruses RRV and SA11-4S reach the cytoplasm from MEs. (g) GTPase Rab7 participates in the formation of LE compartments; ILVs increase in number. (h) The stability and function of LEs depend on the arrival of cellular factors (e.g., cathepsins) from the trans-Golgi network, traffic that is mediated by M6PRs, and the GTPase Rab9, among other factors. (i) L-P RV strains reach late endosomes. RV nar3's exit from LEs requires the function of Rab9. (j) RV strains UK, Wa, WI61, DS-1, and YM require, in addition to Rab9, the function of the CD-M6PR and the activity of cathepsins to productively infect cells. (f, i, j) The cytosolic double-layered particles begin transcribing the RV genome to continue the replication cycle of the virus. The different colors of the viruses represent those strains that exit from MEs (orange) or from LEs that do not require cathepsins (blue) or form LEs, but do require the activity of CD-M6PR and cathepsins (green). HBGA, human histo-blood group antigen; LBPA, lysobisphosphatidic acid.