Figure 10.
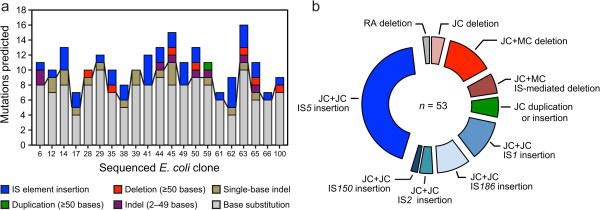
Reanalysis of evolved E. coli samples for structural variation. a) Summary of mutations predicted by breseq in 21 clones sequenced after 6,000 generations of growth in a mutation accumulation experiment [19]. These samples were previously analyzed for single-base substitutions and small indels. The line extending across the bars separates single-base substitutions and indels from mutations affecting more bases that were classified as structural variants. Full details for all mutations predicted in the ancestor of this experiment and each evolved lineage are provided in Additional file 2. b) Overall representation of the different types of structural variants predicted from combinations of new junction (JC) and missing coverage (MC) evidence across all 21 genomes. One structural variant was predicted from spurious read alignment (RA) evidence, as described in the text.
