FIG 3.
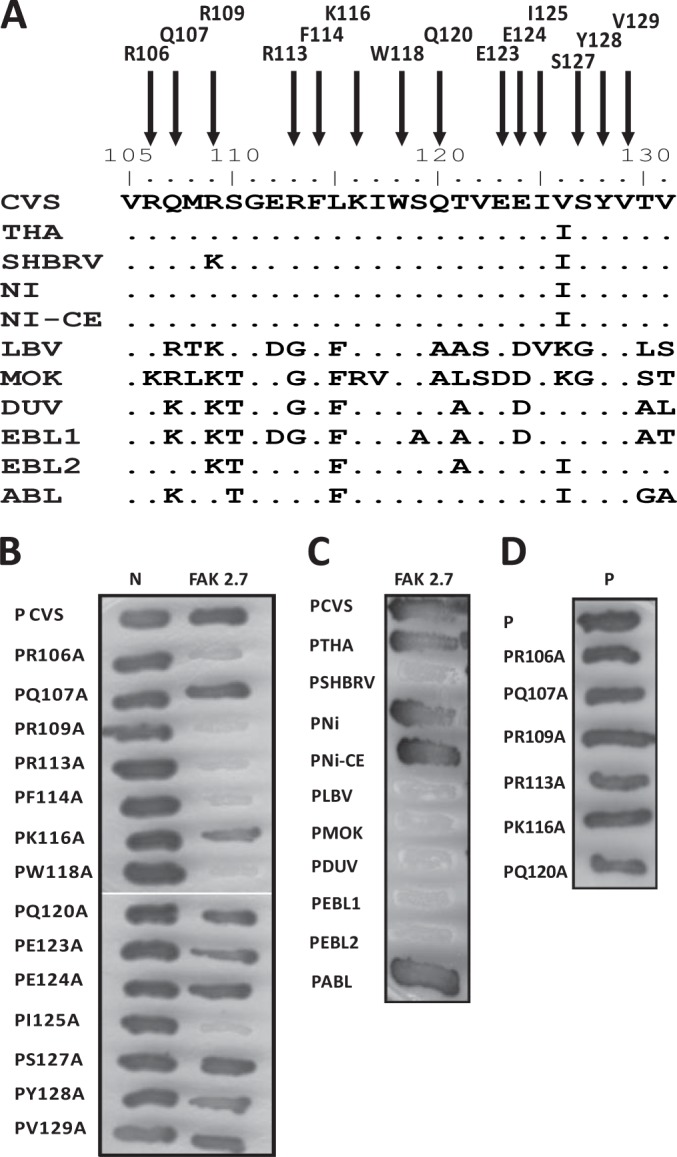
Identification of P residues essential for FAK interaction and analysis of the interaction of FAK with the P proteins of lyssaviruses by two-hybrid screen. (A) Sequence alignment of the FAK interaction domain (residues 106 to 131) of lyssavirus P proteins. Shown are the amino acid sequences of the P proteins of the RABV strains CVS, SHBRV, THA, Ni, and Ni-CE and the lyssaviruses LBV, MOKV, DUVV, EBLV-1, EBLV-2, and ABLV. (B) Binding of mutant P proteins to FAK and RABV N. The interactions between mutant P proteins (in which amino acids have been replaced by an alanine) and FAK (residues 649 to 958) or RABV N were assayed as for Fig. 1A. (C) Binding of FAK to P proteins of lyssaviruses. The interactions were performed as for Fig. 1A. (D) Binding of mutant P proteins to P. The interactions were performed as for Fig. 1A.
