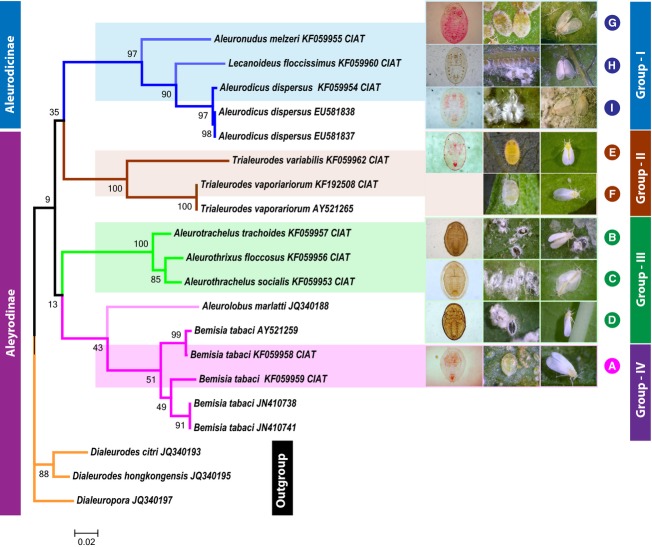
Figure 1.
Phylogenetic relationships inferred using the neighbor-joining method in MEGA V6 and CLC main workbench V6.9 for tropical whitefly species, in relation to well-characterized whitefly taxa for which sequences were available in the GeneBank (Saitou and Nei 1987; Tamura et al. 2011). The optimal tree with the sum of branch length = 1.67299211 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) are shown next to the branches (Felsenstein 1985). The analysis involved 29 amino acid sequences. There were a total of 219 positions in the final dataset. Four well-defined whitefly phylogenetic groups (G-I to G-IV) were observed. The nine species identified at CIAT were named from A to I. A = Aleuronudus melzeri, B = Lecanoideus floccissimus, C = Aleurodicus dispersus, D = Trialeurodes variabilis, E = Trialeurodes vaporariorum, F = Aleurotrachelus trachoides, G = Aleurothrixus floccosus, H = A. trachoides, and I = Bemisia tabaci.