FIG 4.
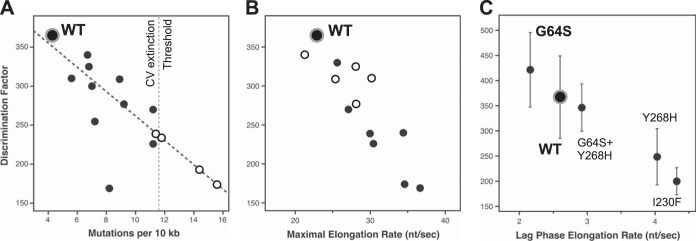
Fidelity modulation of coxsackievirus B3 3Dpol. (A) Correlation plot comparing in vitro coxsackievirus 3Dpol CTP-versus-2′-dCTP discrimination factors with in vivo mutation rates obtained by sequencing of progeny genomes. A linear interpolation of data from viable viruses (black circles) was used to predict in vivo mutation rates of several nonviable variants (open circles) that appeared fully functional in the biochemical assays, leading to the proposed CV extinction threshold of ≈11 mutations per 104 nt (see Table 2 for a complete list of residue identities). (B) Correlation plot comparing the in vitro NTP discrimination factors of CV 3Dpol mutants with their maximal in vitro elongation rates to show that the low-fidelity polymerases have higher elongation rates. Mutants in the palm domain are shown as solid circles, while those in the fingers are shown as open circles. (C) Discrimination factor-versus-rate plot showing that the G64S mutant is a slightly high-fidelity variant of CV 3Dpol based on data obtained at 30°C and pH 7. Several other mutants were also tested at this temperature for direct comparison, as indicated. Note that the x axis is the lag-phase elongation rate observed with nonsaturating 60 μM ATP and 10 μM each GTP and UTP and not the maximal rate from an all-NTP titration as for panel B.
