FIG 4.
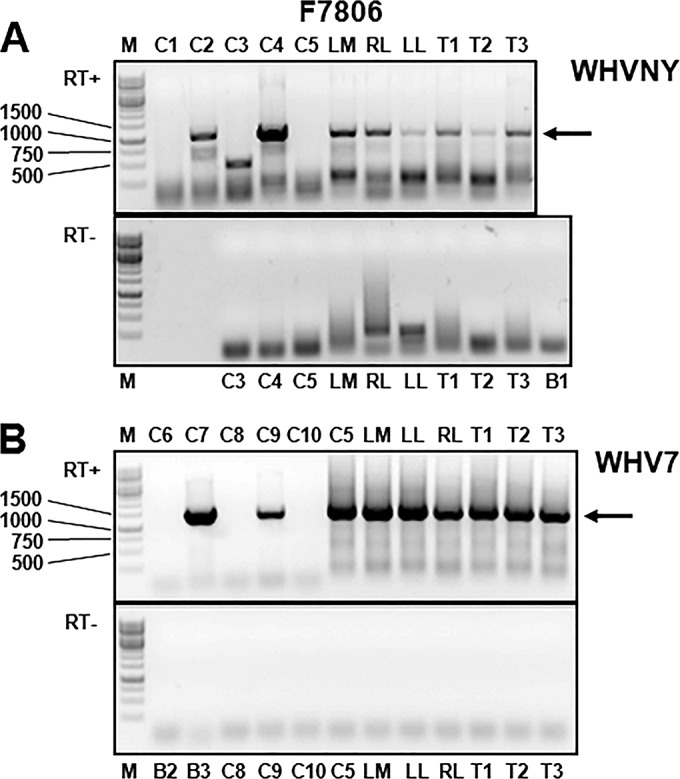
Detection of WHVNY RNA and WHV7 RNA in the samples of nonmalignant liver tissues and matching HCC tissues of woodchuck F7806. The isolation of total RNA and conventional WHV strain-specific PCR assays that amplified either RNA of WHVNY or RNA of WHV7 are described in Materials and Methods. The tissue samples were harvested during necropsy. The PCR products were resolved in 1% agarose gels stained with ethidium bromide. (A) Results of WHVNY-specific PCR assay. (B) Results of WHV7-specific PCR assay. The gel image at the top of each panel demonstrates the results of RNA analysis performed in the presence of reverse transcriptase (RT+). The gel image at the bottom of each panel shows the results obtained in the absence of reverse transcriptase (RT−). The examined tissue samples and the control samples are indicated at the top or bottom of the gel images. The HCC tissues are identified as T numbers, i.e., T1 stands for HCC1, etc. The controls and DNA and RNA standards are the same as those described for Fig. 3. The preparation of WHV strain-specific DNA and RNA control standards is described in Materials and Methods. DNA controls were amplified in the absence of RT. WHVNY RNA was detected in all assayed tissue samples during at least two independent RNA isolations/examinations (Table 1). The specific PCR products of the expected sizes (see the legend to Fig. 3 for details) are indicated with arrows on the right. The DNA size markers are shown on the left (lanes labeled M).
