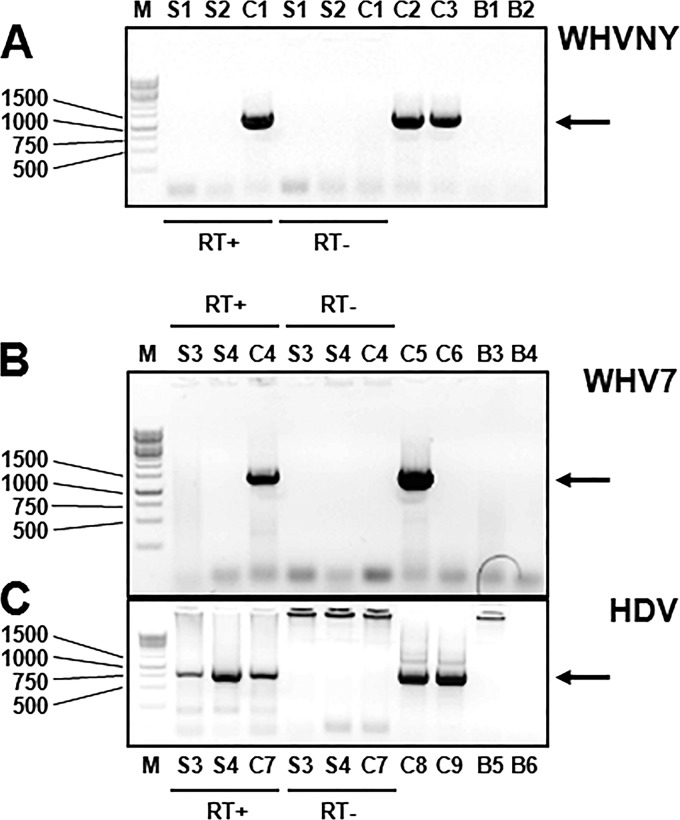
FIG 6.
Experimental proof that WHV RNA was not detected in the sera of woodchucks that were either transiently or chronically infected with WHV. The conventional PCR assays that amplified exclusively either WHVNY RNA or WHV7 RNA are the same as the ones that were used in experiments presented in Fig. 3 to 5. The HDV-specific nested PCR assay that amplifies the genomic HDV RNA, which is present in HDV virions, is detailed in Materials and Methods. In all panels, the reactions performed in the presence of the reverse transcriptase are labeled RT+, and the reactions performed in the absence of the reverse transcriptase are marked RT−. The details regarding WHV strain-specific RNA and DNA control standards are described in Materials and Methods. The PCR products were examined in 1% agarose gels stained with ethidium bromide. (A) Analysis of total RNA isolated from the serum samples of woodchucks F6678 and F6541 using WHVNY-specific PCR assay. For RNA isolation, serum collected from animal F6678 at week eight after monoinfection with the strain WHVNY and serum from animal F6541 harvested at week 11 after monoinfection with WHVNY were used. The samples that were assayed were the following: S1, total RNA from the serum of F6678; S2, total RNA from serum of F6541; C1, 5.0 GE of WHVNY RNA standard; C2, 20 GE of WHVNY DNA standard; C3, 2.0 GE of WHVNY DNA standard; B1, no template in the 1st PCR; B2, no template in the 2nd PCR. Panels B and C show the results of the analysis of total RNA isolated from the sera collected from two woodchucks, F6438 and M6593, which were chronic carriers of WHV7 that also were superinfected with WHV-enveloped hepatitis delta virus (wHDV). A serum sample from woodchuck F6438 was collected at 4 weeks after wHDV superinfection, while serum from M6593 was harvested at 5 weeks after wHDV superinfection. (B) Analysis conducted using WHV7-specific nested PCR assay. (C) Results of the analysis that used HDV-specific nested PCR assay. The samples that have been assayed were the following: S3, total RNA isolated from serum of F6438; S4, total RNA from serum of M6593; C4, 5.0 GE of WHV7 RNA standard; C5, 20 GE of WHV7 DNA standard; C6, 2.0 GE of WHV7 DNA standard; C7, 5.0 × 103 GE of HDV genomic RNA standard (prepared as described previously [31]); C8, 3.7 × 1010 HDV DNA standard (plasmid pSVLD3, which harbors the trimer of the HDV genome [51]); C9, 3.7 × 109 HDV DNA standard; B3, no template in the 1st PCR; B4, no template in 2nd PCR; B5, no template in the 1st PCR; B6, no template in 2nd PCR. The DNA control standards were amplified in the absence of RT. The specific PCR products of the expected sizes are indicated with arrows on the right. The expected sizes of WHVNY RNA-derived and WHV7 RNA-derived PCR products are described in the legend to Fig. 3. The expected size of the HDV-specific PCR product was 819 bp. The DNA size markers are shown on the left (lanes labeled M).