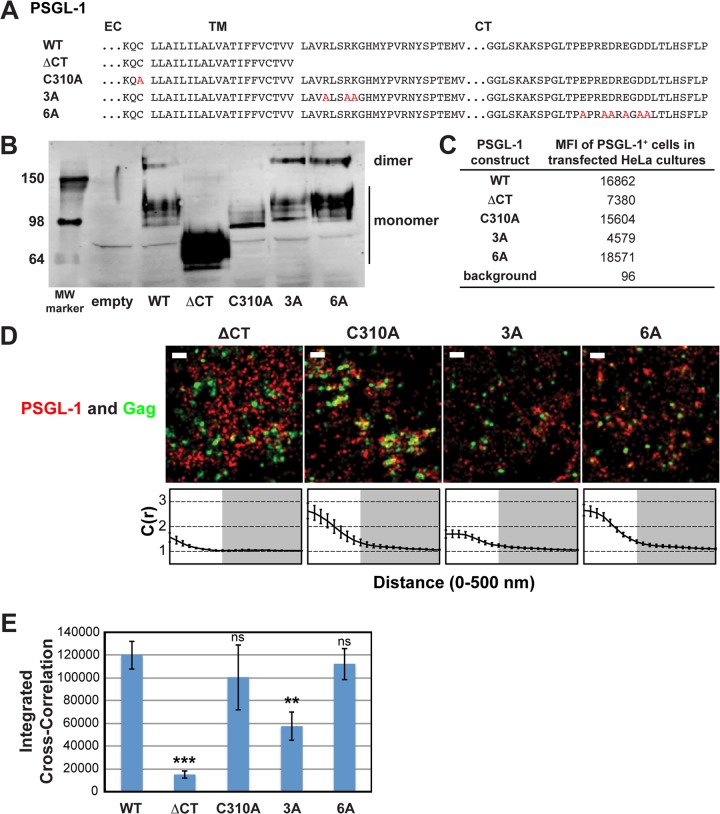
FIG 3.
A basic sequence in the cytoplasmic tail of PSGL-1 enhances coclustering with HIV-1 Gag. (A) Partial amino acid sequences of WT and mutant PSGL-1 constructs. EC, extracellular domain; TM, transmembrane domain; CT, cytoplasmic tail. Amino acid substitutions are shown in red. (B) WT PSGL-1 and PSGL-1 mutants were expressed in HeLa cells, and cell lysates were analyzed by SDS-PAGE (nonreducing conditions), followed by immunoblotting using an anti-PSGL-1 antibody. Numbers on the left are molecular weights (MWs), in kilodaltons. (C) HeLa cells transfected with plasmids carrying the indicated PSGL-1 constructs or an empty vector were recovered as cell suspensions at 16 h posttransfection, fixed, immunostained with an anti-PSGL-1 antibody labeled with Alexa Fluor 647, and analyzed by flow cytometry. The background mean fluorescence intensity was determined using cells transfected with the empty vector. Cells expressing transfected PSGL-1 derivatives, which were readily distinguished on the basis of the much higher fluorescence intensity, were gated, and their mean fluorescence intensities (MFIs) were determined. (D) HeLa cells were transfected with plasmids carrying WT Gag-mEos3.2 and the indicated PSGL-1 constructs and imaged as described in the legend to Fig. 2B. Representative reconstructed images are shown. Bars = 500 nm. Gag-mEos3.2 is shown in green, and PSGL-1 is shown in red. Cross-correlation curves, prepared as described in the legend to Fig. 1A, are shown. (E) The analysis of total coclustering was performed as described in the legend to Fig. 1B using cross-correlation curves for images of a total of 10 cells per condition from 2 independent experiments. Values shown indicate means ± SEMs. P values were calculated for each construct relative to WT PSGL-1. ***, P < 0.0005; **, P < 0.005; ns, not significant.